Healthy Control Human iPSC Line, Male, SCTi004-A
Human pluripotent stem cell line, frozen

Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration
Overview
To ensure optimal product performance and reproducibility, SCTi004-A is manufactured using extensive quality control procedures in a culture system consisting of mTeSR™ Plus, Corning® Matrigel® hESC-Qualified Matrix, and ReLeSR™. SCTi004-A is karyotypically stable, demonstrates trilineage differentiation potential, expresses undifferentiated cell markers, and was reprogrammed using a non-integrating reprogramming technology. Registration with hPSCreg® ensures ethical and biological conformity based on community standards.
SCTi004-A has been validated with STEMdiff™ kits across multiple lineages and tissue types in both 2D and organoid models (Figures 6-11). Choose TeSR™ and STEMdiff™ cell culture media products to establish a complete workflow for your cell culture system. For alternative genotypes for your iPSC control lines, see all of our high-quality, healthy control human iPSC lines, female and male.
This research-use-only (RUO) product has been consented for both academic and commercial use. Blood samples are ethically sourced using Institutional Review Board (IRB) or other regulatory authority-approved consent forms and protocols. For donor details and cell quality characterization of the source cell banks, refer to the data figures on this page. For additional details, refer to the lot-specific Certificate of Analysis and Frequently Asked Questions about induced pluripotent stem cell lines.
Whole exome and whole genome sequence data files are available upon request. Please contact us for pricing.
Certain products are only available in select territories. Please contact your sales representative or Product & Scientific Support at techsupport@stemcell.com for further information.
Data Figures
Table 1. iPSC Line SCTi004-A Is Derived from a Healthy Male Donor
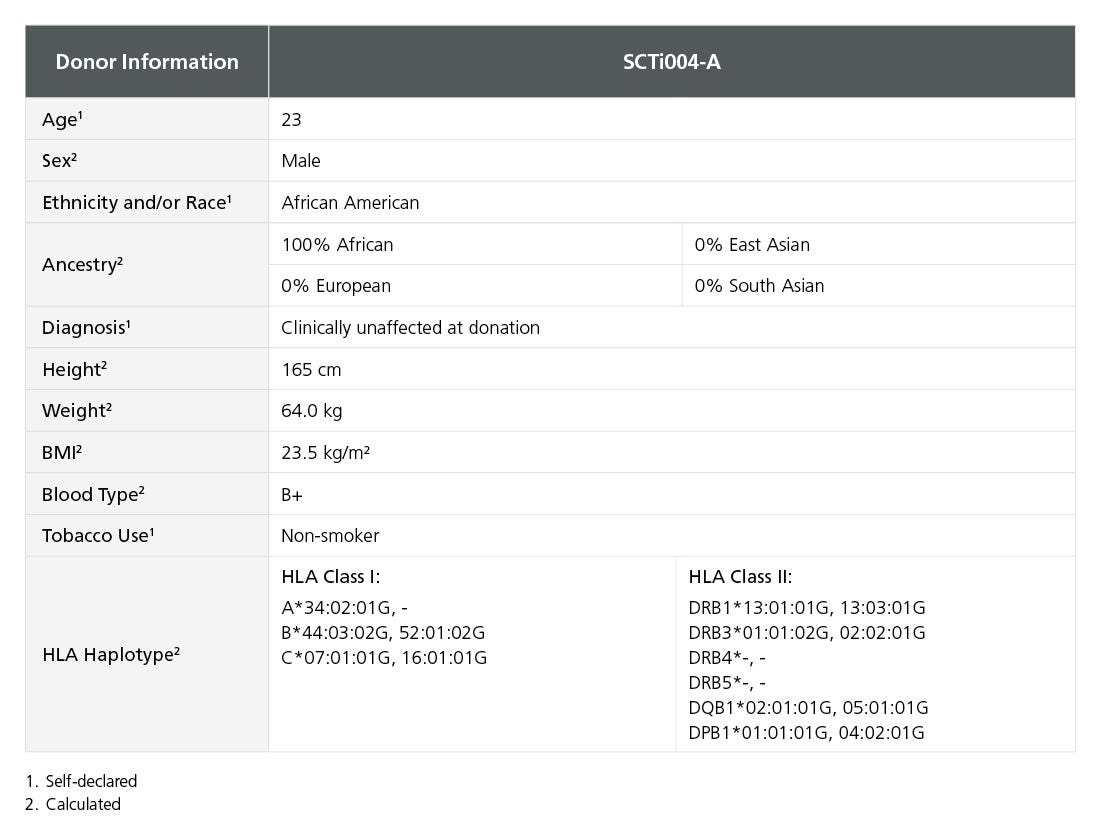
Demographic, health, and genetic characteristics of the SCTi004-A donor are compiled based on self-reported information and whole-exome sequencing. Sex was determined by karyotype. Ancestry was calculated by EthSEQ analysis from whole-exome sequencing data. HLA haplotype was determined by next-generation sequencing, sequence-base typing, and sequence-specific oligonucleotide probes as needed to obtain the required resolution. Blood type (ABO/Rh blood group) was determined by next-generation sequencing. Height, weight, and BMI were calculated at the donation facility. iPSC = induced pluripotent stem cell.
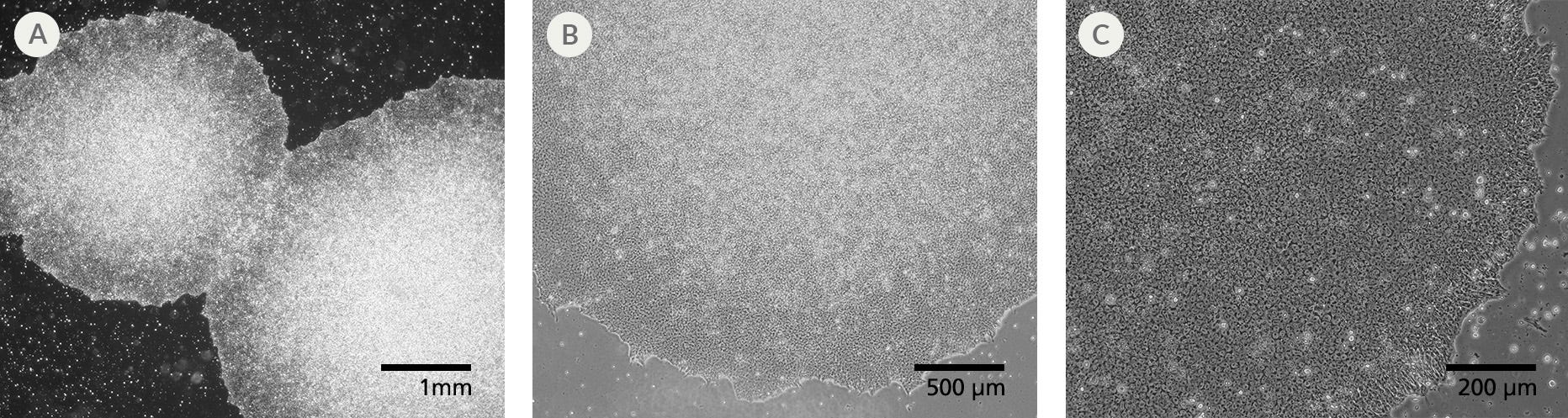
Figure 1. SCTi004-A Human Pluripotent Stem Cells Demonstrate High-Quality Morphology in Routine Culture
Cryopreserved cells from line SCTi004-A were thawed and maintained in mTeSR™ Plus on Corning® Matrigel® Matrix. (A) The resulting iPSC colonies have densely packed cells and show multi-layering when ready to be passaged. (B,C) Cells retain prominent nucleoli and high nuclear-to-cytoplasmic ratios. iPSC = induced pluripotent stem cell.
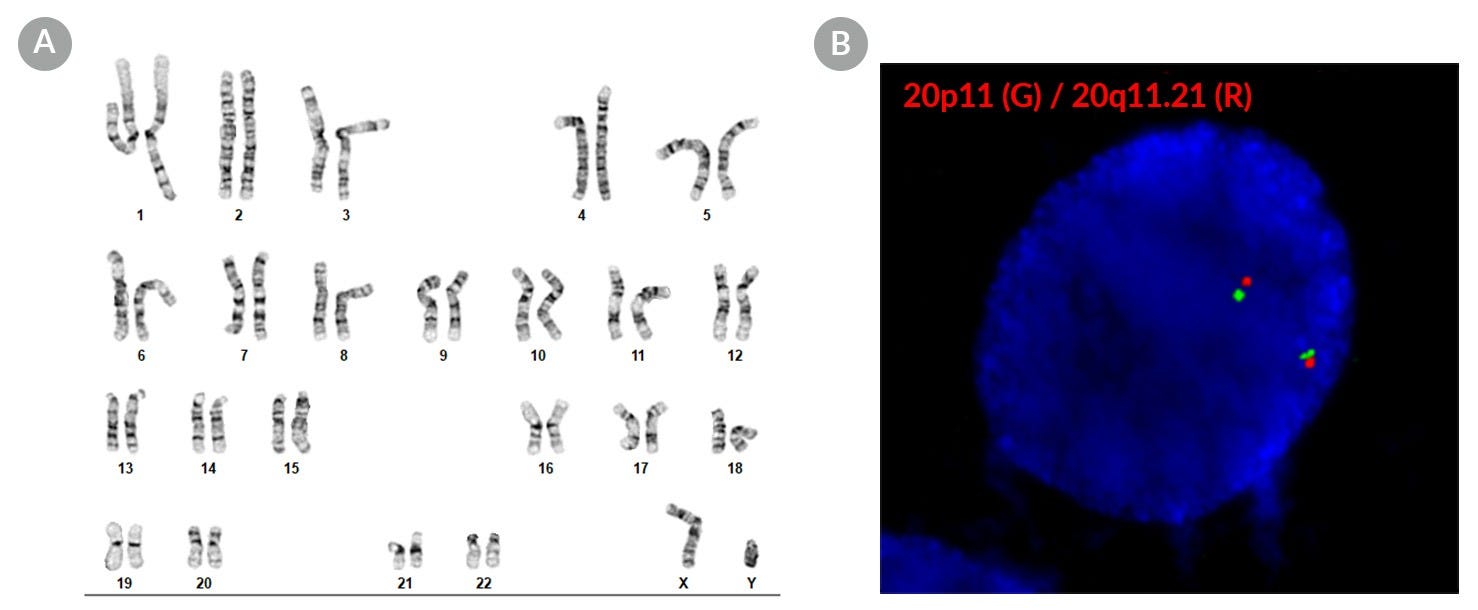
Figure 2. SCTi004-A Human Pluripotent Stem Cells Maintain a Normal Karyotype
(A) G-T-L banding for thawed cells at p20 (n = 20) shows a normal karyotype with no evidence of clonal abnormalities at a band resolution of 450 - 550 G-bands per haploid genome. (B) Fluorescent in situ hybridization in a representative p20 iPSC using probes for 20p11 (green) and 20q11.21 (red). 94% of cells examined displayed two sets of two probe signals, indicating no aneusomy of chromosome 20 (n = 200). iPSC = induced pluripotent stem cell.
Table 2. Single Nucleotide Polymorphism Microarray Analysis Characterizes SCTi004-A Copy Number Variants
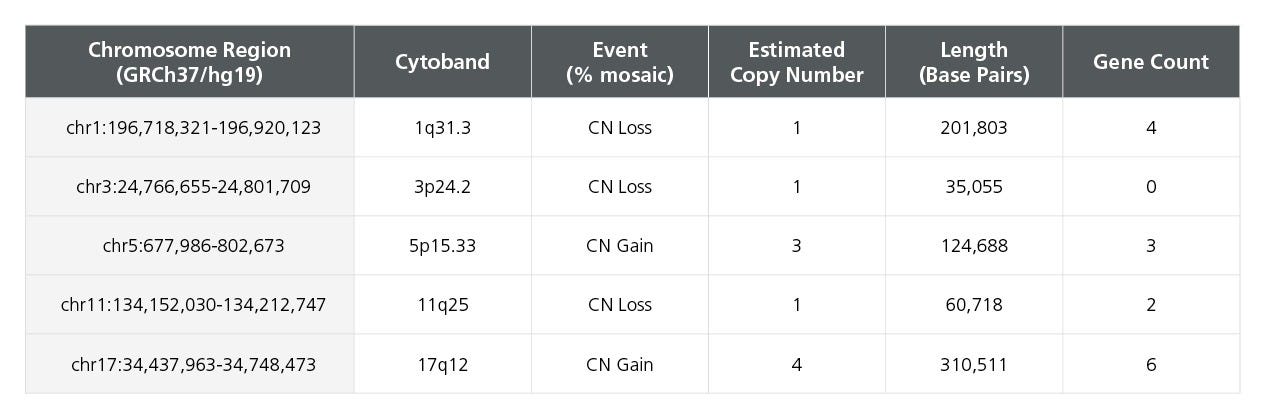
DNA was extracted from a vial of SCTi004-A and subject to SNP microarray analysis to identify large-scale copy number variants (CNVs). The cells display no reportable CNVs, defined as those greater than 400kb in size. Array design, genomics position, genes, and chromosome banding are based on genome build GRCh37/hg19. chr = chromosome; start cyto = cytogenetic band at the start of the base pair imbalance; end cyto = cytogenetic band at the end of the base pair imbalance; bp = base pairs; SNP = single nucleotide polymorphism.
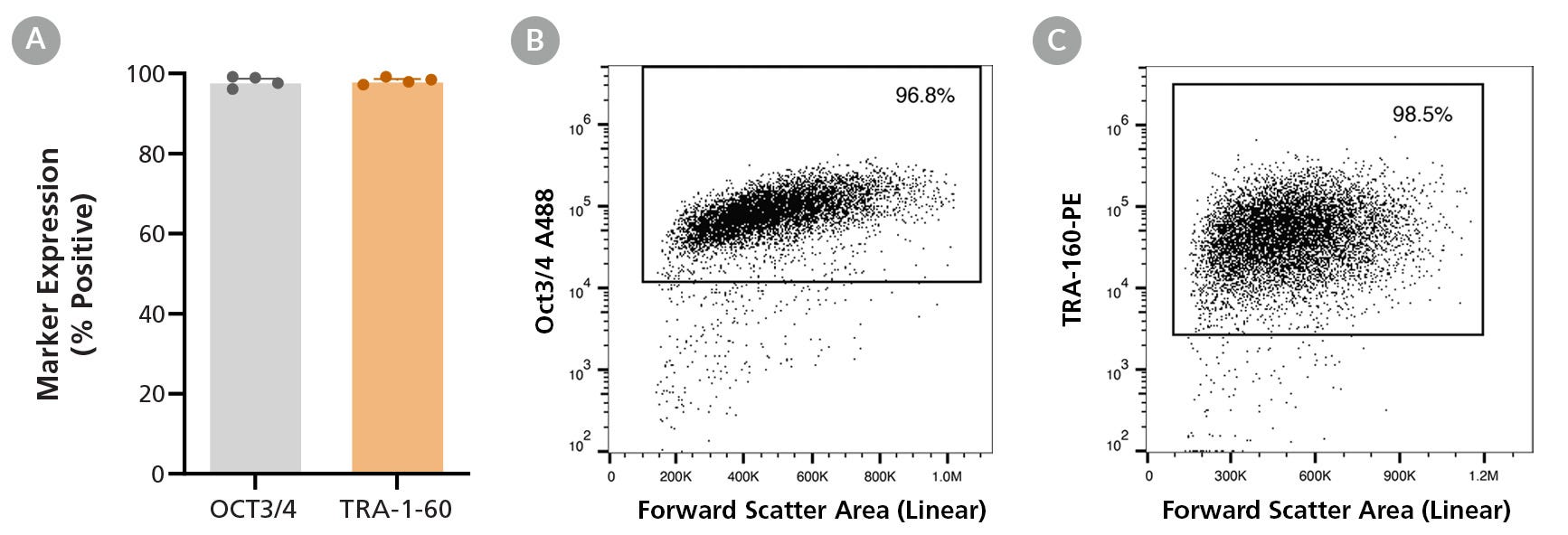
Figure 3. SCTi004-A iPSCs Express Undifferentiated Cell Markers
Cell line SCTi004-A was characterized using flow cytometry for undifferentiated cell markers OCT3/4 and TRA-1-60. (A) Percentage marker expression was quantified 5 passages after thawing from the analyses of four technical replicates. Representative flow cytometry plots are displayed for (B) TRA-1-60 and (C) OCT3/4. iPSC = induced pluripotent stem cell.
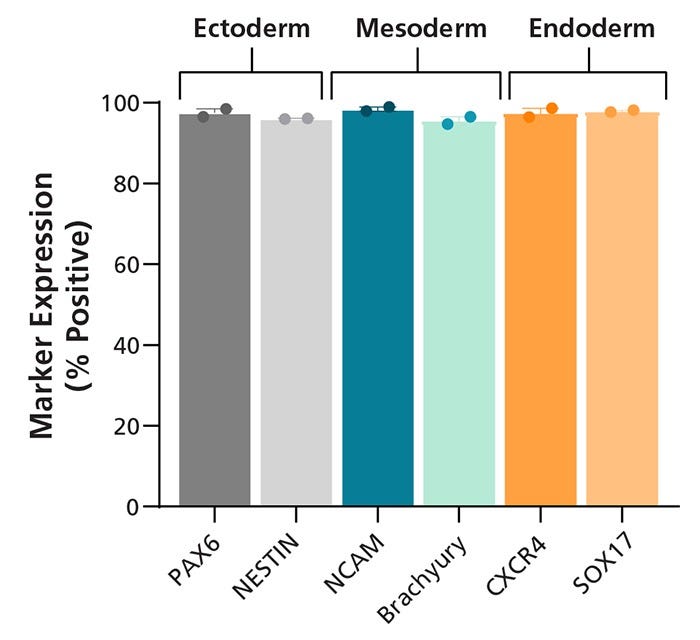
Figure 4. SCTi004-A Human Pluripotent Stem Cells Demonstrate a High Trilineage Differentiation Capacity
Cells from SCTi004-A were split into 3 groups, differentiated using STEMdiff™ Trilineage Differentiation Kit (Catalog #05230), and then subjected to flow cytometry analysis. Two markers for each embryonic germ layer were assessed, and bars present mean marker expression for each group of cells (n = 2 biological replicates). PAX6 and Nestin confirm differentiation to the ectoderm lineage, NCAM and Brachyury (T) to the mesoderm lineage, and CXCR4 and SOX17 to the endoderm lineage.
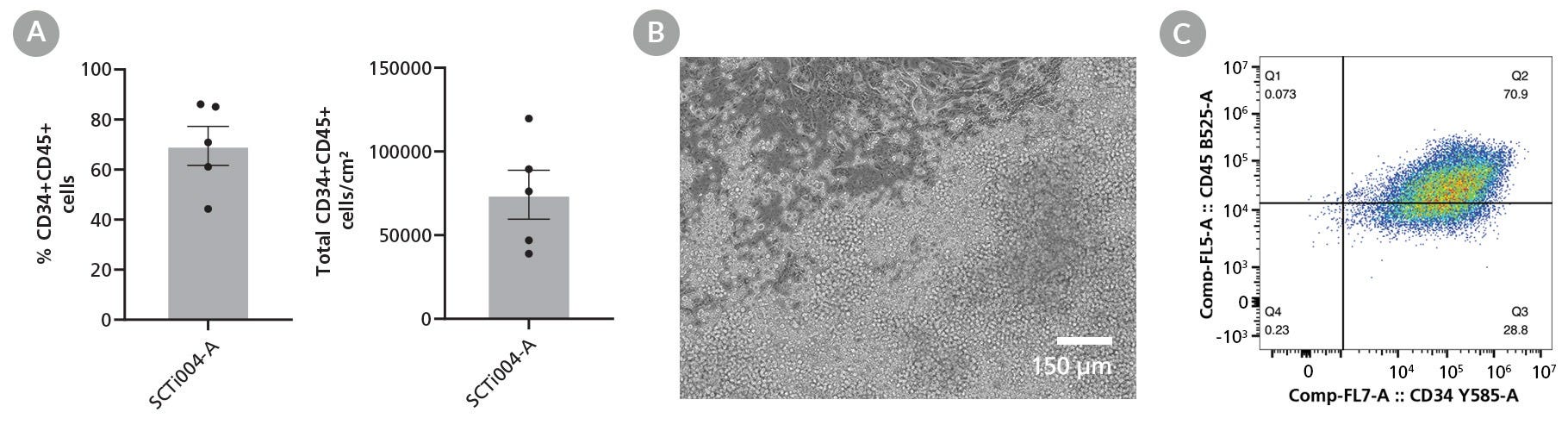
Figure 5. SCTi004-A iPSCs Can Form Hematopoietic Progenitor Cells (HPCs)
STEMdiff™ Hematopoietic Kit (Catalog #05310) was used to generate HPCs from the SCTi004-A cell line. (A) Percentages and yields of CD34+CD45+ HPCs per cm2 after differentiation. Error bars represent standard error (n = 5 biological replicates). (B) Brightfield image of SCTi004-A-derived HPCs indicates that these cells transitioned through a typical endothelial-to-hematopoietic transition (EHT).
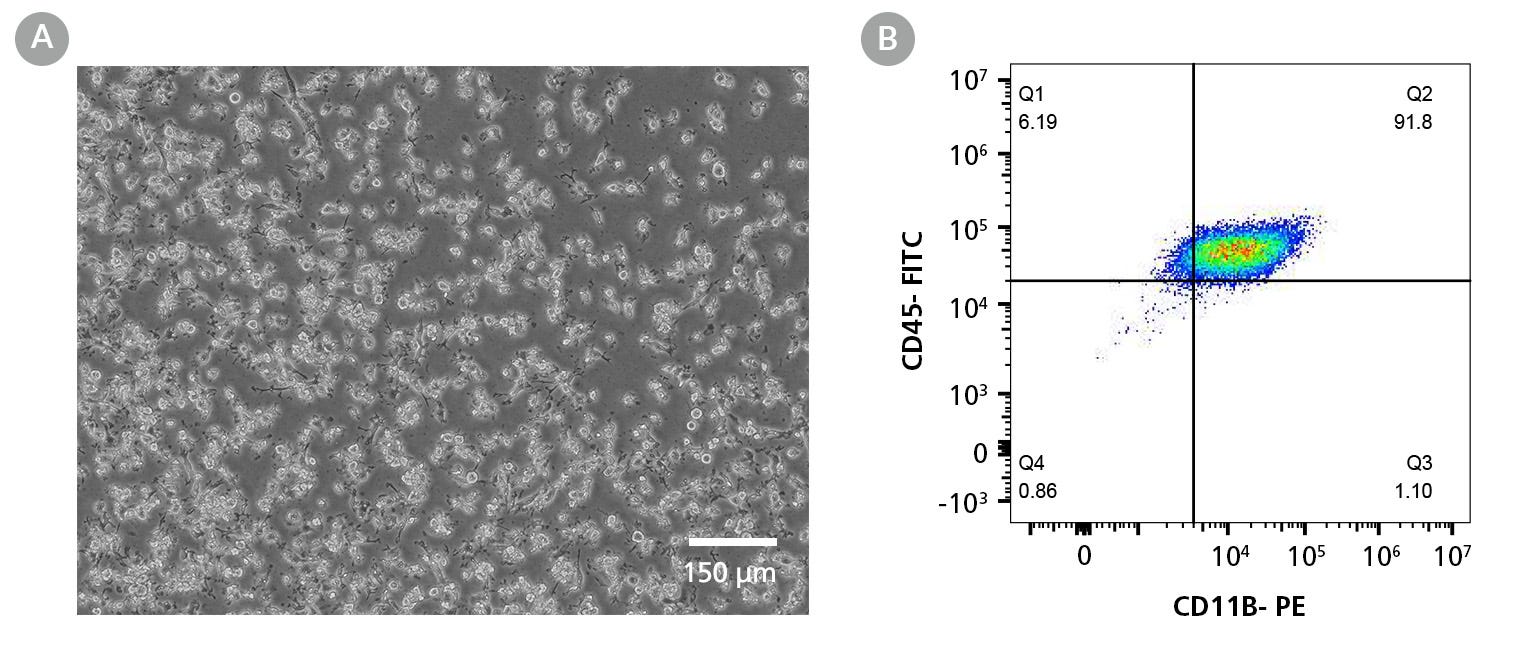
Figure 6. SCTi004-A Human Pluripotent Stem Cells Can Effectively Differentiate into Microglia
Hematopoietic progenitor cells generated from cell line SCTi004-A using STEMdiff™ Hematopoietic Kit (Catalog #05310) were further differentiated using STEMdiff™ Microglia Differentiation and Maturation Kits (Catalog #100-0019, #100-0020). (A) The resulting cells are small with visible processes, are non-adherent on Matrigel®, and exhibit small cytoplasmic-to-nuclear ratios characteristic of microglia. (B) Co-expression of CD45 and CD11b was observed by flow cytometry.
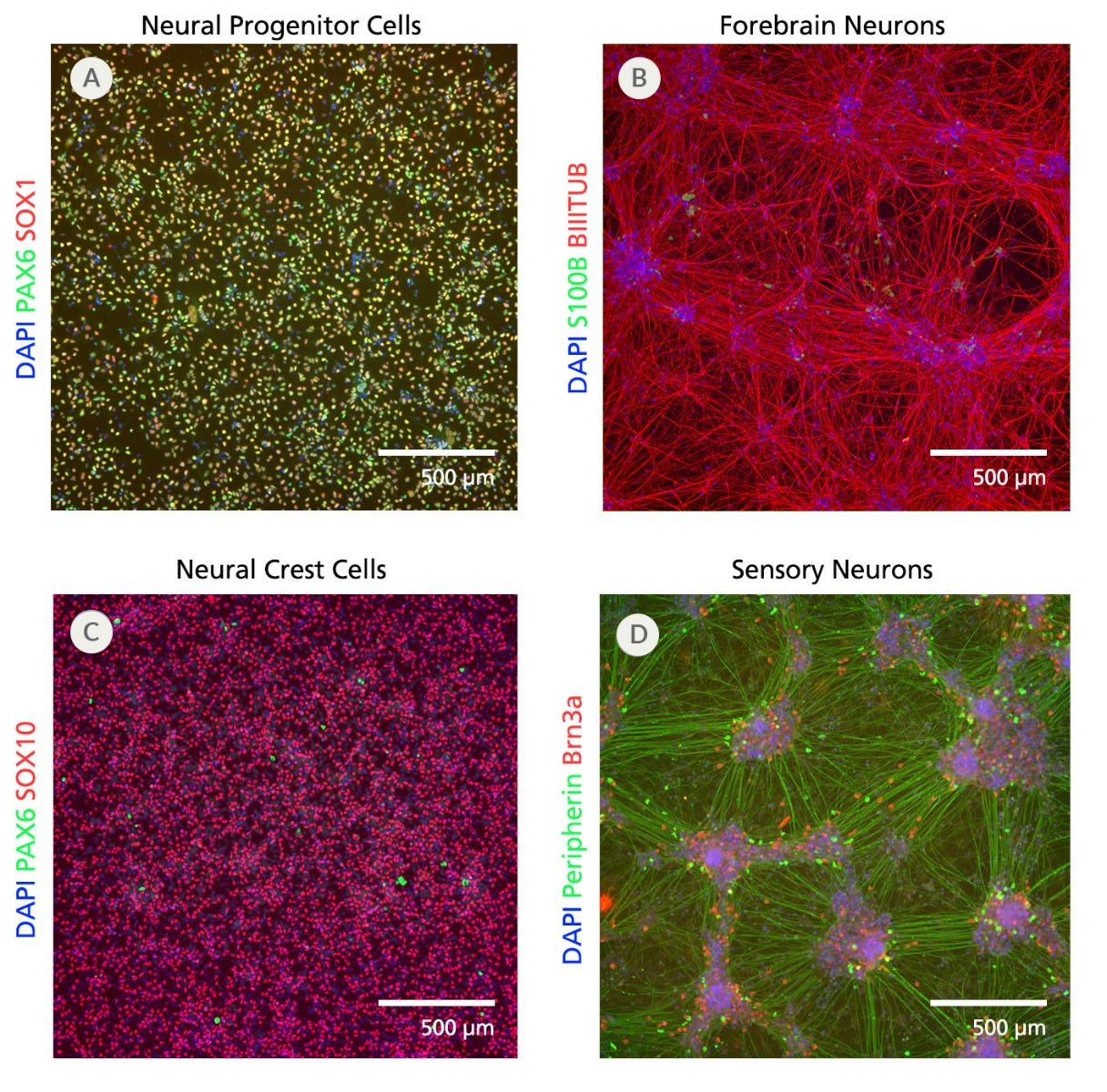
Figure 7. SCTi004-A Human Pluripotent Stem Cells Can Efficiently Differentiate into Neural Progenitor Cells, Forebrain Neurons, Neural Crest Cells, and Sensory Neurons
(A) SCTi004-A iPSCs were differentiated into (A) neural progenitor cells using STEMdiff™ SMADi Neural Induction Kit (Catalog #08581), (B) forebrain neurons using STEMdiff™ Forebrain Neuron Differentiation Kit (Catalog #08600), (C) neural crest cells using STEMdiff™ Neural Crest Differentiation Kit (Catalog #08610), and (D) sensory neurons using STEMdiff™ Sensory Neuron Differentiation Kit (Catalog #100-0341). See the respective product information sheets for more information and the protocols used.
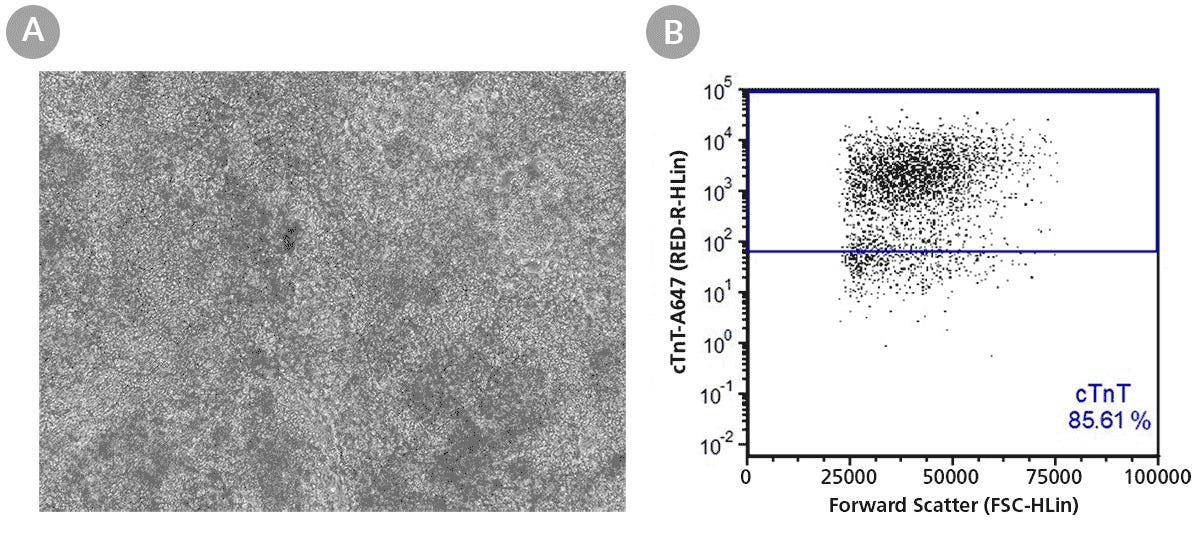
Figure 8. SCTi004-A Human Pluripotent Stem Cells Can Successfully Differentiate into Ventricular Cardiomyocytes
Ventricular cardiomyocytes were generated from SCTi004-A iPSCs using STEMdiff™ Ventricular Cardiomyocyte Differentiation Kit (Catalog #05010). (A) Monolayer cultures at Day 15 of differentiation show iPSC-derived ventricular cardiomyocytes that exhibit beating behavior. (B) Of the resulting cells, 86% express cardiomyocyte marker cTnT, as detected by flow cytometry.
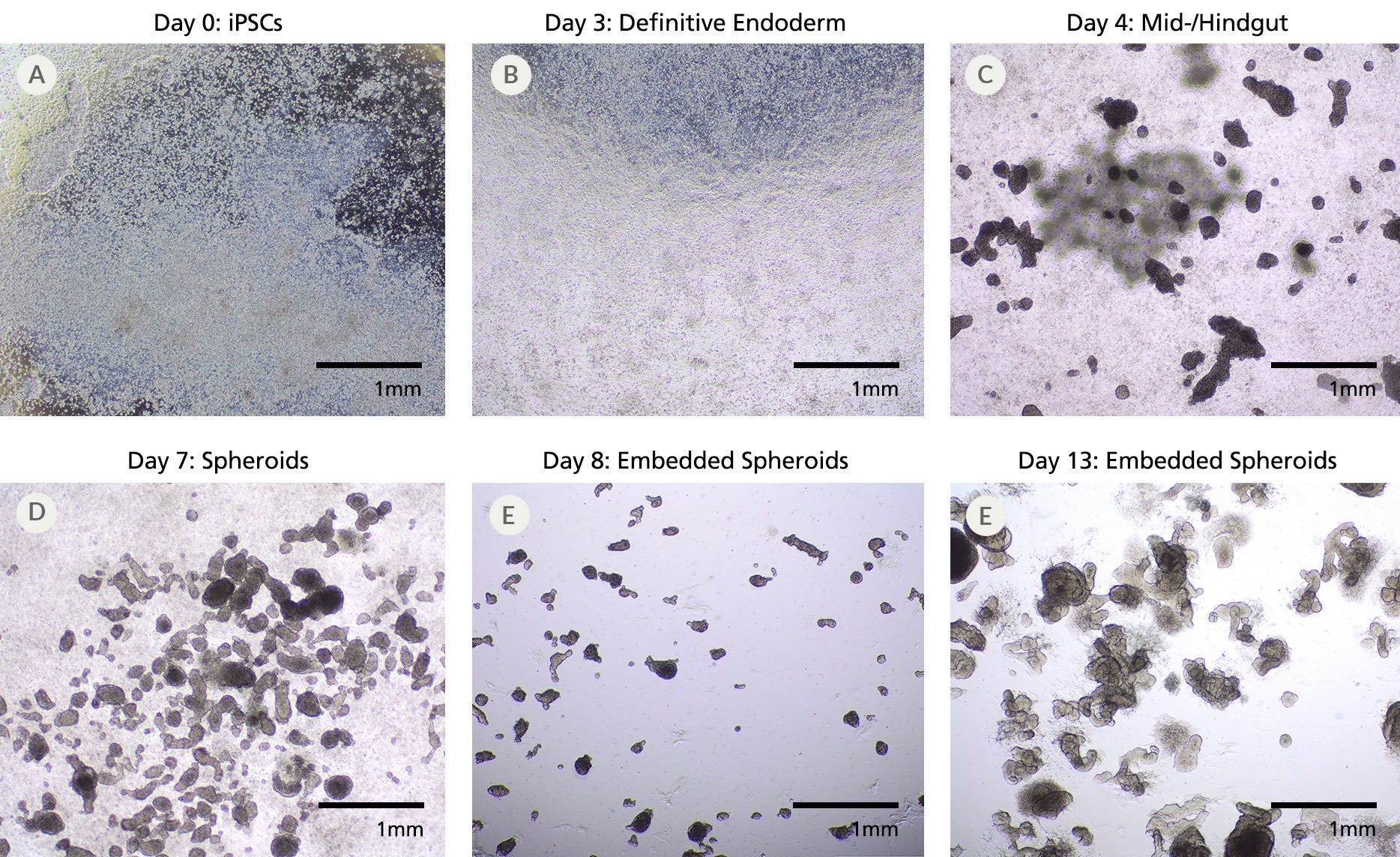
Figure 9. SCTi004-A Human Pluripotent Stem Cells Can Successfully Differentiate into Intestinal Organoids
(A) SCTi004-A iPSCs were seeded for differentiation with STEMdiff™ Intestinal Organoid Kit (Catalog #05140). (B) By Day 3, the monolayers show more uniformity and display characteristics of patterning to definitive endoderm. (C) After switching to mid-/hindgut medium, 3D structures become visible atop the monolayer culture, and (D) spheroids detach from the mid-/hindgut culture at Day 7 of differentiation. (E) Collections of spheroids can be embedded in Matrigel® domes for subsequent maturation into human intestinal organoids, which (F) expand significantly after just 6 days in matrix. The maturing organoids can be passaged and expanded using STEMdiff™ Intestinal Organoid Growth Medium (Catalog #05145). iPSCs = induced pluripotent stem cells.
Table 3. Comprehensive Risk Variant Profile for the SCTi004-A iPSC Line
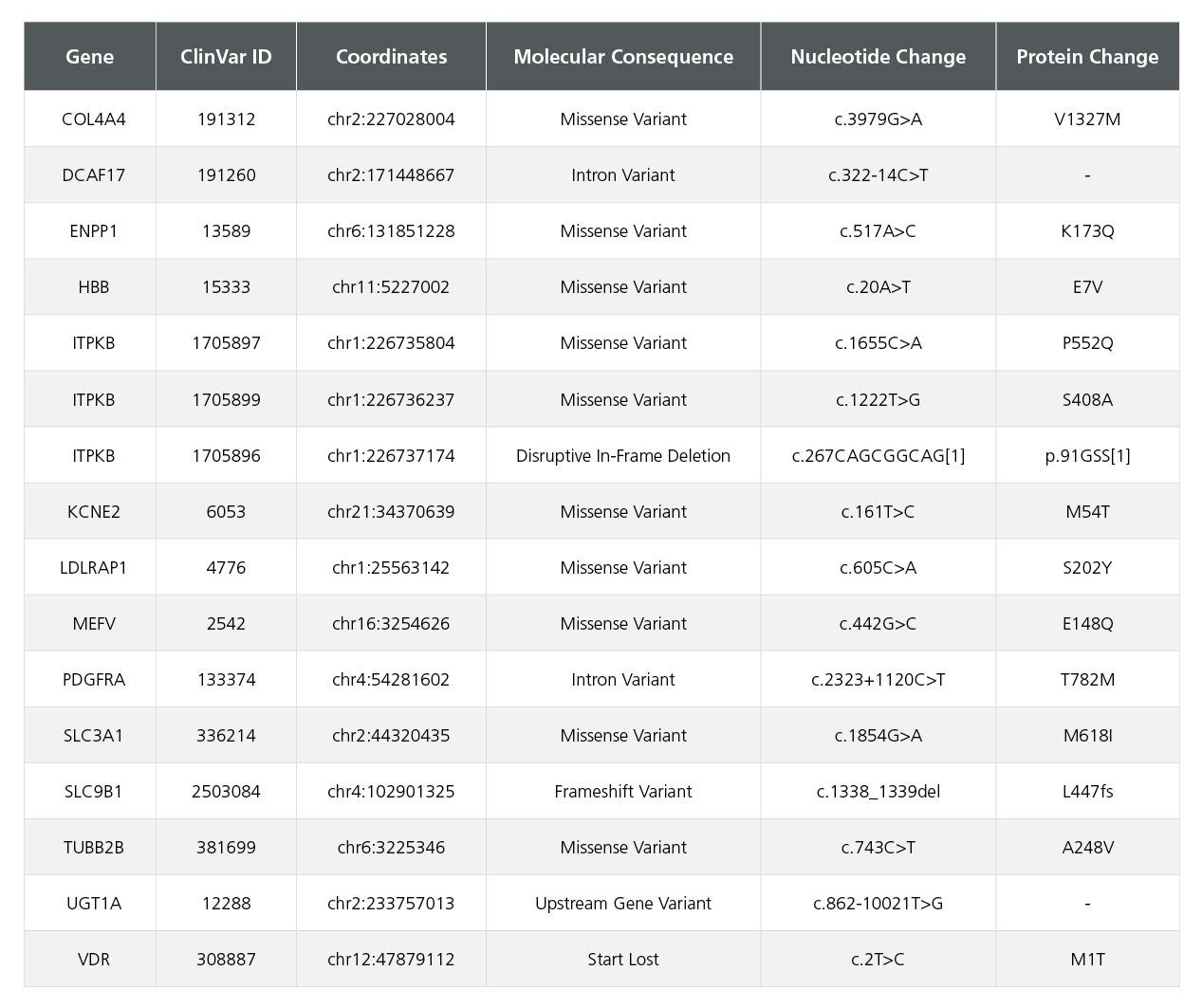
The table presents risk variants identified for the SCTi004-A iPSC line. DNA was purified from the master cell bank, and whole-genome sequencing was performed to capture the entire genomic landscape. Sequencing was conducted on the NovaSeq 6000 System, achieving a 50x coverage. Variants were called and analyzed following GATK4 best practices, incorporating a Convolutional Neural Network to score each variant. High-confidence variants were annotated with predicted functional consequences and ClinVar IDs. ClinVar entries required two or more supporting ALT reads and an assertion criteria of one gold star or more in ClinVar to be considered. This comprehensive profile assists in understanding the genetic landscape and potential health impacts associated with the SCTi004-A iPSC line.
Protocols and Documentation
Find supporting information and directions for use in the Product Information Sheet or explore additional protocols below.
Applications
This product is designed for use in the following research area(s) as part of the highlighted workflow stage(s). Explore these workflows to learn more about the other products we offer to support each research area.
Resources and Publications
Educational Materials (34)
Related Products
-
 ReLeSR™
ReLeSR™cGMP, enzyme-free human pluripotent stem cell selection and passaging reagent
-
 mTeSR™ Plus
mTeSR™ PluscGMP, stabilized feeder-free maintenance medium for human ES and iPS cells
-
 CloneR™2
CloneR™2Defined supplement for improving survival of human ES and iPS cells in single-cell workflows
-
 Healthy Control Human iPSC Line, Female, SCTi...
Healthy Control Human iPSC Line, Female, SCTi...Human pluripotent stem cell line, frozen
-
 CellAdhere™ Laminin-521
CellAdhere™ Laminin-521Matrix for maintenance of human ES and iPS cells in combination with TeSR™ maintenance media
Item added to your cart

Healthy Control Human iPSC Line, Male, SCTi004-A
LIMITED USE LICENSE These iPSCs and their modifications (including but not limited to derivatives or differentiated progeny) shall not be used or administered in (1) human subjects for human clinical use; (2) animals for veterinary use for therapeutic, diagnostic, or prophylactic purposes or (3) any subject in relation to clinical applications, cell therapy, transplantation, and/or regenerative medicines, without limiting the generality of the foregoing. These iPSCs and their modifications (including but not limited to derivatives or differentiated progeny) may not be used for monetization or commercialization purposes, including without limitation, used to, or with the goal to, perform services or supply products or rights, including in the manufacture of cellular therapies or other therapeutics, for monetary gain or the generation of royalties, revenues, sales or other valuable consideration. For clarity, these iPSCs and their modifications (including but not limited to derivatives or differentiated progeny) may not be used for screening compounds, antibodies, proteins or peptides, except for the purposes of target discovery, target validation, or assay development, provided such activities and the results of such activities are not further used for monetization or commercialization purposes. It may be possible to obtain a further license for the prohibited uses referred to in this Limited Use License. Please contact iPSCrequests@stemcell.com for more details.
UNLESS OTHERWISE STATED, PRODUCTS ARE FOR RESEARCH USE ONLY AND NOT INTENDED FOR HUMAN OR ANIMAL DIAGNOSTIC OR THERAPEUTIC USES. FOR PRODUCT-SPECIFIC COMPLIANCE AND INTENDED USE INFORMATION, REFER TO THE PRODUCT INFORMATION SHEET. GENERAL INFORMATION ON QUALITY AT STEMCELL MAY BE FOUND AT WWW.STEMCELL.COM/COMPLIANCE.


































