Improve the Efficiency and Quality of Your Cell Sorting by Pre-Enriching Cells Prior to FACS
Isolating rare cell types by flow sorting alone can be time consuming. Starting your sort with pre-enriched populations allows you to obtain your rare cells faster than by using fluorescence-activated cell sorting (FACS) alone.
- Document # 27137
- Version 2.0.0
- May 2021
In this document, we highlight a method to improve pre-FACS purity and maintain high recoveries of complex immune cell subsets and rare cells, while drastically reducing downstream cell subset sorting time.
Background
Using fluorescence-activated cell sorting (FACS) alone to isolate large numbers of complex or rare cell types can be time-consuming, expensive, and lead to reduced viability of sorted cells. Starting your FACS with a cell population that has been pre-enriched for your cells of interest enables you to more quickly and efficiently isolate complex or rare immune cell subsets than can be achieved using FACS alone, helping you accomplish your research goals faster.
EasySep™, a column-free immunomagnetic cell isolation technology, is one of the fastest and easiest ways to pre-enrich cells prior to FACS. With both negative and positive selection protocols, you can perform upfront bulk removal of unwanted cells from your sample in as little as 8 minutes. The case studies below outline how pre-enrichment of dendritic and innate lymphoid cells using EasySep™ can result in improved purity and higher recovery, thereby drastically reducing cell subset sorting times.
Why Pre-Enrich Cells Before FACS?
- Obtain viable and functional cells that are immediately ready for FACS.
- Increase sample throughput by reducing downstream FACS time.
- Increase the purity of complex or rare cell types.
Case Study: Reducing FACS Time for Dendritic Cell Subset Isolation
Dendritic cells (DCs) are a group of innate immune cells that recognize pathogen-associated molecular patterns (PAMPs) and respond by processing associated extracellular and intracellular proteins to present as antigens in the context of MHC molecules. Through antigen presentation to naïve T cells, DCs bridge the innate and adaptive immune systems. DCs can be further characterized into two separate subsets—myeloid or conventional DCs (cDCs) and plasmacytoid DCs (pDCs).1
Some immune cell subsets, such as cDCs and pDCs, are found at a low frequency in samples. Isolating these cells can require the design of complex multicolor flow cytometry experiments where multiple cell surface markers are fluorescently labeled and used for FACS. Due to their complexity, these FACS experiments can often take hours, which can adversely affect the quality of the sorted cells. Not only can these lengthy experiments directly cause reduced cell viability, but the presence of significant numbers of dead cells in your sample can increase autoflourescence and non-specific binding to fluorescently labeled antibodies, thereby altering scatter properties and compromising the purity of your isolated cells.2,3 Starting your FACS with a pre-enriched cell population increases the frequency of your target cells, helping you achieve faster isolation of complex cell types and reducing the risk of FACS-related cell death that could impact your downstream experiments. Here, we describe a method for pre-enriching a pan-DC population that improves the pre-FACS purity of cDCs and pDCs while maintaining high cell recoveries, thereby drastically reducing the time required for downstream FACS.
Methods
To create a single-cell suspension, mouse splenocytes were digested at 37°C using Spleen Dissociation Medium (Catalog #07915). Single-cell suspensions were first pre-enriched for pan-DCs using immunomagnetic negative selection with EasySep™ Mouse Pan-DC Enrichment Kit II (Catalog #19863) prior to staining with fluorescently labeled antibodies. DC subsets were then analyzed using flow cytometry. Alternatively, red blood cells were first lysed prior to staining and flow cytometry. Lineage-negative markers were used to identify DCs (CD3e, CD19, Ly-6G, F4/80, NK1.1 (CD161), TER119, and IgM) by flow cytometry. cDCs were defined as Lin-CD11c+ PDCA-1- and pDCs were defined as Lin-CD11clowPDCA-1+. Final purities were compared between the two isolation methods, and the percentage recoveries of cDCs and pDCs were calculated in EasySep™-enriched samples. To calculate protocol times, each sample was run on the flow cytometer for an equivalent amount of time with a consistent flow rate and the data was used to extrapolate the total time required to sort 1 million pan-DCs, cDCs, or pDCs using FACS.
Results and Summary
Starting with a single-cell suspension of mouse splenocytes, pre-enrichment with EasySep™ Mouse Pan-DC Enrichment Kit II increased the pre-FACS purity of cDCs from 2.9% to 44.9% and pDCs from 0.8% to 7.1%, and resulted in high recoveries of both cell subsets for downstream FACS (Figures 1).
To evaluate the effect of pre-enrichment on the time required to obtain DC subsets using FACS, unenriched and EasySep™-enriched samples were run on the flow cytometer and the data was used to extrapolate the total theoretical time required to sort 1 million DCs and DC subsets using FACS. Pre-enrichment with EasySep™ Mouse Pan-DC Enrichment Kit II reduced the time to sort 1 million pan-DCs by 79.6%, cDCs by 80.2%, and pDCs by 87.1% (Figure 2). On average, the overall time to acquire cDCs was reduced from 3.5 hours to just 42 minutes, and pDCs from 11.5 hours to just 91 minutes, saving significant time.
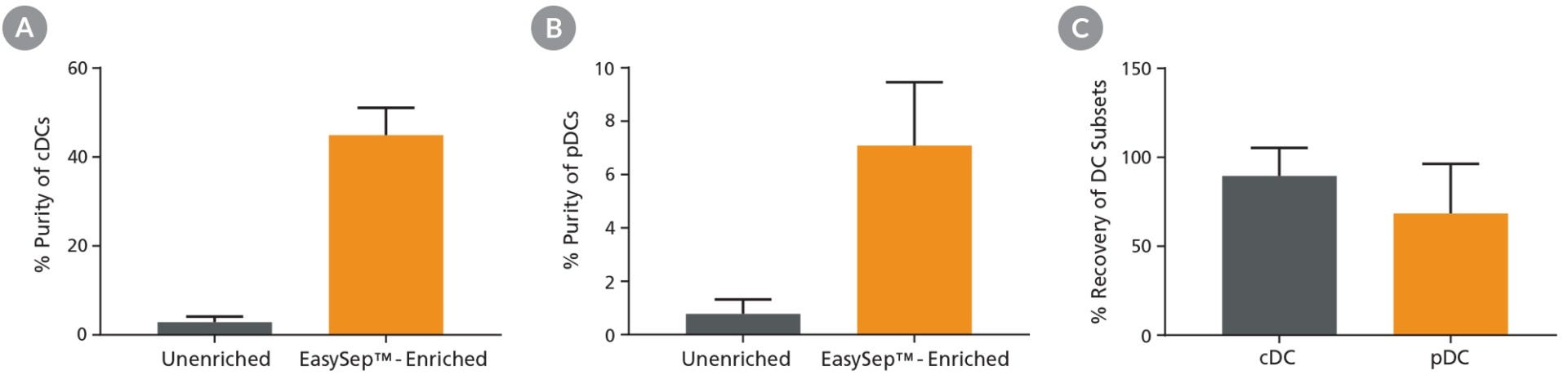
Figure 1. Pre-Enrichment Results in Improved Purity and High Recovery of DC Subsets
Following mouse splenocyte digestion and red blood cell lysis, unenriched samples were stained with fluorescently labeled antibodies for flow cytometric analysis of conventional DCs (cDCs) or plasmacytoid DCs (pDCs). Alternatively, pan-DCs were first enriched using EasySep™ prior to being run on the flow cytometer. Cell purity of (A) cDCs and (B) pDCs was measured using flow cytometry in unenriched and EasySep™-enriched samples. (C) The % recovery of DC subsets was calculated for EasySep™-enriched samples. Data shown as mean ± SD; n = 5 - 9.
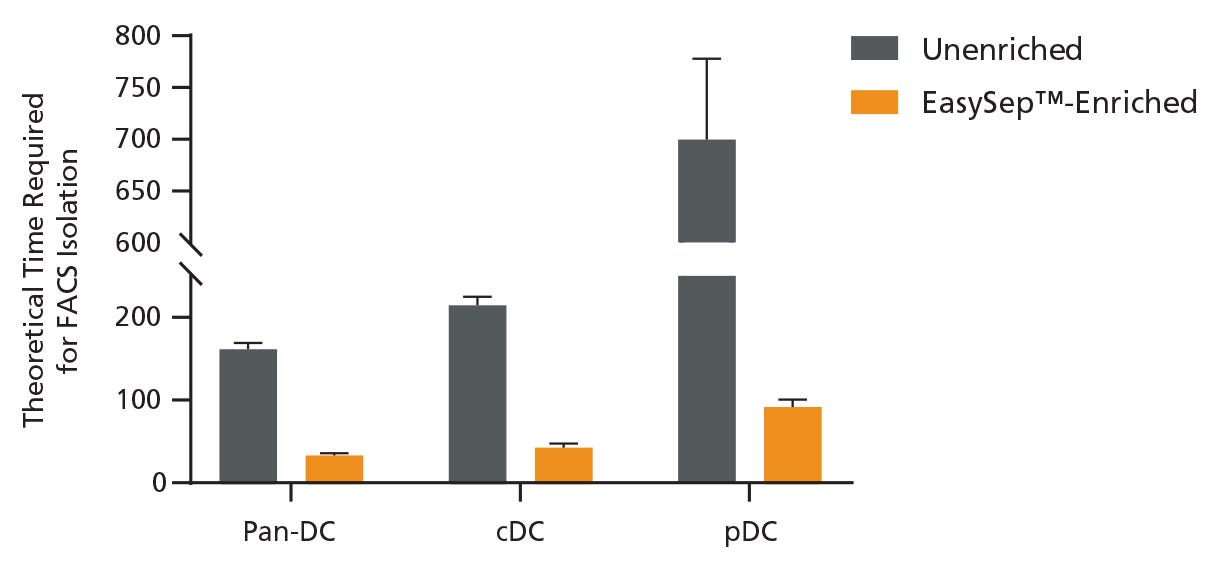
Figure 2. Pre-Enrichment Reduces the Time Required to Obtain DC Subsets by FACS
Starting with a single-cell suspension of mouse splenocytes, pan-DCs, conventional DCs (cDCs), and plasmacytoid DCs (pDCs) were analyzed in parallel from unenriched or EasySep™-enriched samples by flow cytometry. In the unenriched sample, red blood cells were first lysed before performing flow cytometry. Alternatively, pan-DCs were first enriched using EasySep™ before analysis on the flow cytometer. Each sample was run on the flow cytometer for an equivalent amount of time with a consistent flow rate and the data was used to extrapolate the total theoretical time required to sort 1 million pan-DCs, cDCs, or pDCs using FACS. Data shown as mean ± SD; n = 2.
Case Study: Improving the Isolation and Analysis of Rare ILC Subsets
Innate lymphoid cells (ILCs) are important effector cells in innate immunity and have essential functions in immune regulation, inflammation, and tissue homeostasis. In healthy individuals, ILCs are extremely rare, comprising <0.1% of CD45+ leukocytes in mouse and human peripheral blood.4,5 The low frequency and lack of unique surface markers for ILCs create challenges when it comes to their identification, isolation, and characterization. ILCs are typically isolated using multicolor FACS; however, this method is time-consuming and expensive, and can result in low cell recovery. Here, we discuss a method to drastically reduce the time required to isolate rare ILC subsets (ILC1, ILC2, and ILC3) while maintaining the functionality of the isolated cells.
Methods
Single-cell suspensions were prepared by washing unprocessed leukapheresis samples twice with phosphate-buffered saline containing 2% fetal bovine serum. Following staining of the single-cell suspensions with fluorescently labeled antibodies, ILCs were isolated by FACS. Alternatively, single-cell suspensions were pre-enriched for target cells using EasySep™ Human Pan-ILC Enrichment Kit (Catalog #17975) prior to staining and FACS. ILCs were identified as CD45+ , lineage-negative (CD1a, CD3, CD11c, CD14, CD19, CD34, CD123, TCRαβ, TCRγδ, BDCA-2, FcεR1α, CD94, CD4, CD16), and CD127+ by flow cytometry. ILC1s were further defined as CD117- CRTH2- , ILC2s as CD117+ CRTH2+ , and ILC3s as CD117+ CRTH2- . Final purities, cytokine production, and protocol times were compared between the two isolation methods.
Results and Summary
Starting with 7.5 x 107 total cells and a pre-FACS purity of 0.1%, isolation by FACS alone resulted in a final ILC purity of 74% and took approximately 120 minutes of FACS time. A second round of FACS was required in order to obtain a final purity of 97% (data not shown). Extrapolating from these results, it would take approximately 3,200 minutes of FACS time to process 2 x 109 cells (Figure 3). Conversely, starting with 2 x 109 total cells from the same donor, pre-enrichment using EasySep™ increased the pre-FACS ILC frequency from 0.1% to 27%. FACS time for this EasySep™-enriched sample was reduced to 12 minutes, with a final Lin− CD127+ cell purity of 99% (Figure 3). Sorted ILC1, ILC2, and ILC3 subsets from EasySep™-enriched samples remained functional as assessed by cytokine secretion (Figure 4).
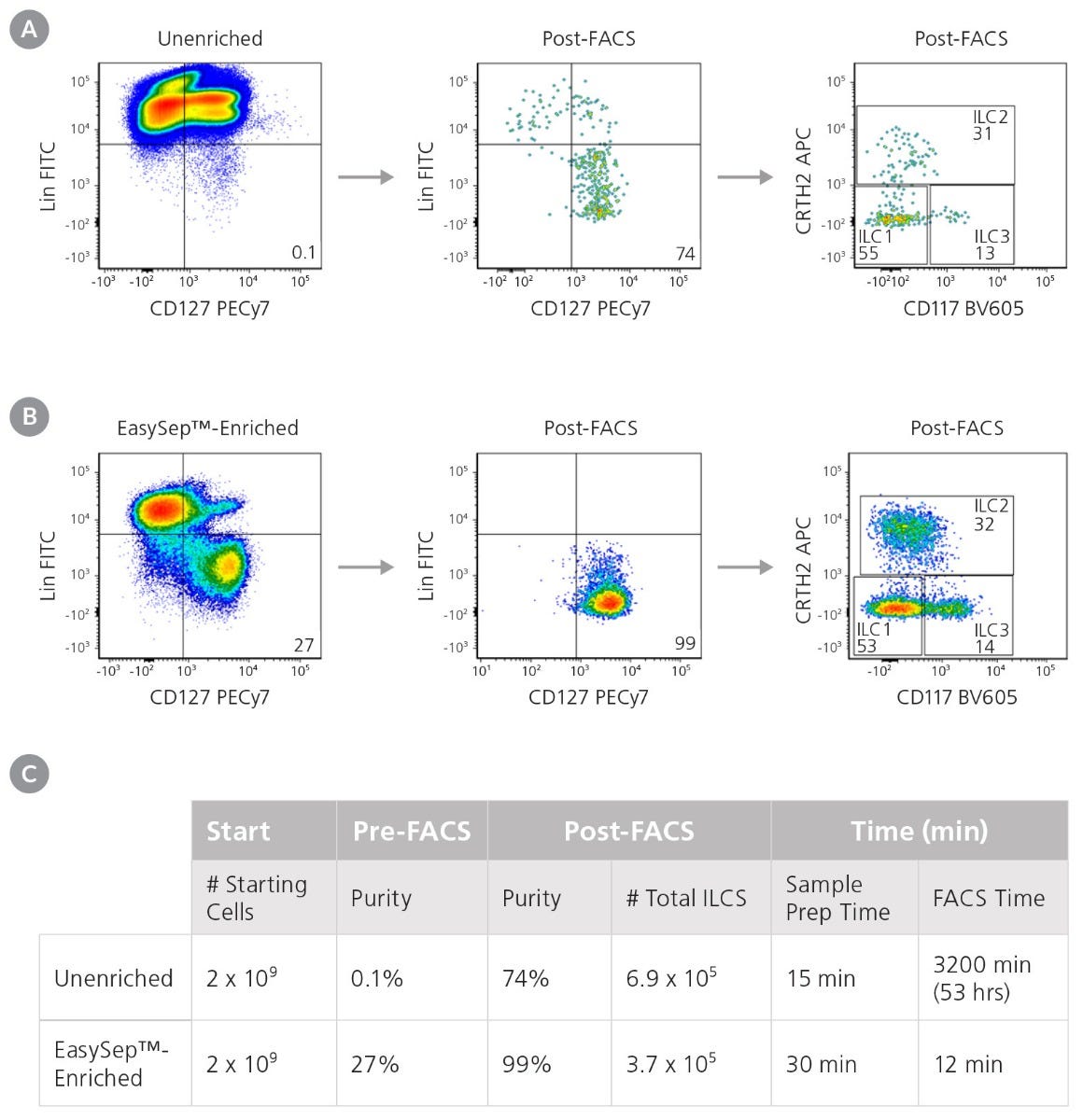
Figure 3. Pre-Enrichment with EasySep™ Reduces the Time Required to Obtain Pure ILCs
Starting with a fresh leukapheresis sample, ILCs s (Lin-CD127+) were isolated in parallel from an unenriched or an EasySep™-enriched sample. (A) In an unenriched sample, ILC frequency was assessed by flow cytometry at the start and after one round of FACS. (B) In an EasySep™-enriched sample, ILC frequency was assessed immediately after EasySep™-enrichment, and again after one round of FACS. (C) Corresponding purities and FACS times at each stage are reported.
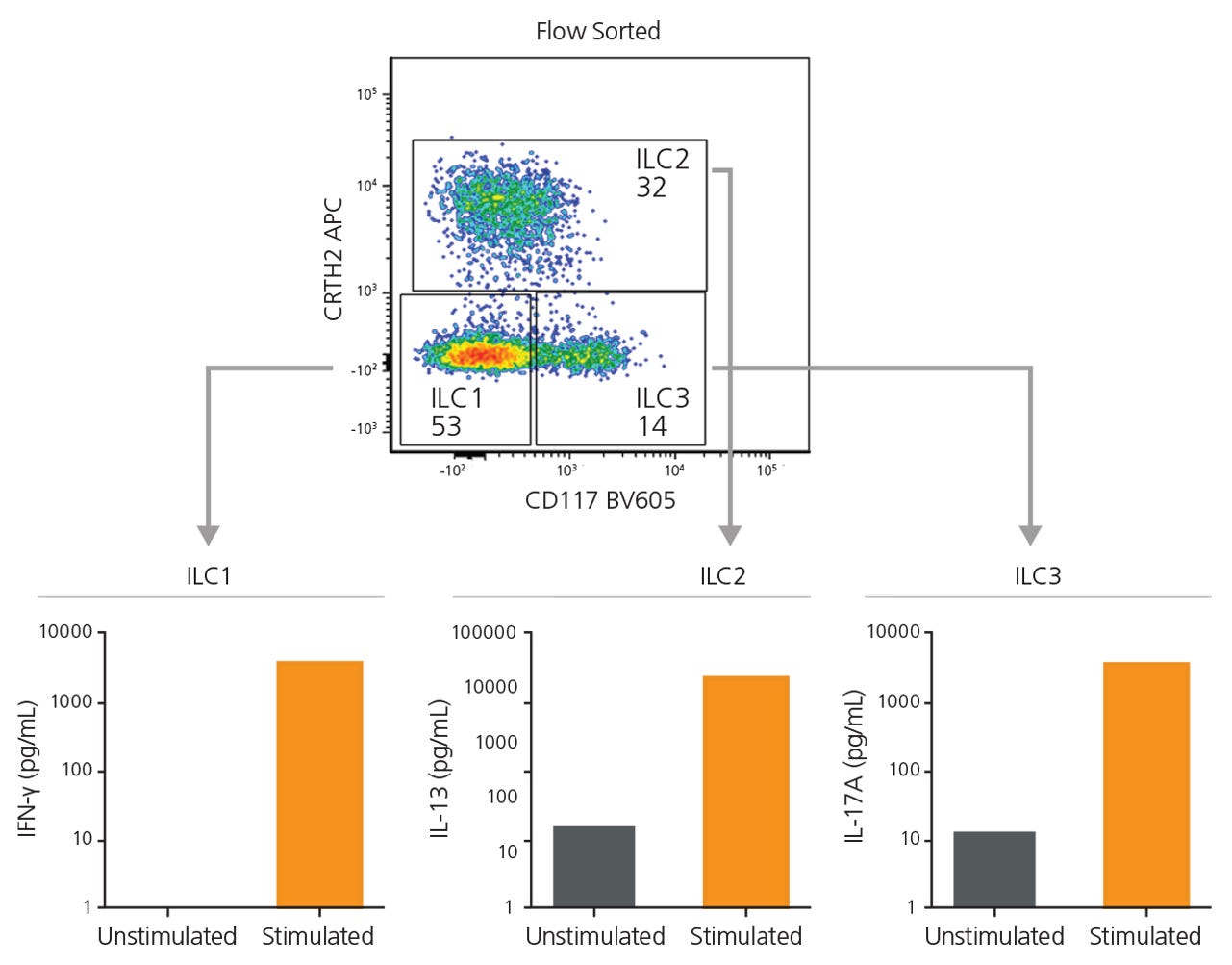
Figure 4. EasySep™-Enriched ILCs Are Functional
ILC1, ILC2, and ILC3s sorted from EasySep™-enriched samples were cultured independently in a serum-free expansion medium, with or without stimuli. After 6 days, supernatants from ILC1 (IL-12 and IL-15), ILC2 (IL-2 and IL-33), and ILC3 (IL-2, IL-1β and IL-23) cultures were collected and analyzed for the secretion of IFNγ, IL-13, and IL-17A, respectively, by enzyme-linked immunosorbent assay.
Conclusion
Isolating complex immune cell subsets or rare cells by FACS can be time-consuming and can negatively impact the purity of your isolated cells, slowing you down in reaching your research goals. In the case studies above, we show how EasySep™ can be used to quickly deplete unwanted cells to enrich for rare, untouched, and functional DC subsets and ILCs that are ready for downstream analysis or FACS. Overall, adding EasySep™ upstream of FACS can result in drastically reduced FACS times while improving the quality of your isolated cells.
Products for Cell Pre-Enrichment Prior to FACS
EasySep™ is a column-free immunomagnetic cell separation technology that enables rapid isolation of highly purified and functional cells for downstream applications. Pre-enrichment of cell populations using EasySep™ reduces FACS time and enables isolation of complex immune cell subsets and rare cell types.
Table 1. Examples of Reagents for Human Cell Enrichment and Downstream Subset Sorting
EasySep™ Human CD4+ T Cell Isolation Kit (17952)
EasySep™ Human CD8+ T Cell Isolation Kit (17953)
Th2 Cells
Th17 Cells
Th9 Cells
Naïve T Cells
Central Memory T Cells (Tcm)
Effector Memory T Cells (Tem)
T Follicular Helper Cells (Tfh)
CD161+ Regulatory T Cells (Treg)
Naïve Tregs
Invariant Natural Killer T Cells (iNKT)
Exhausted T Cells (PD1high)
Effector T Cells
Plasmacytoid DCs (pDC)
Table 2. Examples of Reagents for Mouse Cell Enrichment and Downstream Subset Sorting
EasySep™ Mouse CD4+ T Cell Isolation Kit (19852)
EasySep™ Mouse CD8+ T Cell Isolation Kit (19853)
Memory T Cells (Tmem)
Regulatory T Cells (Treg)
Gamma-Delta T Cells
Natural Killer T Cells (NKT)
Antigen-Specific T Cells
Naïve T Cells (CD45RBhigh)
Plasma Cells
Plasmacytoid DCs (pDC)
ILC2
ILC3
Polymorphonuclear MDSCs (PMN-MDSC)
EasySep™ Mouse CD117 (cKIT) Positive Selection Kit (18757)
EasySep™ Mouse Hematopoietic Progenitor Cell Isolation Kit (19856)
Long-Term Hematopoietic Stem Cells (LT-HSC)
Short-Term Hematopoietic Stem Cells (ST-HSC)
Explore More Technical Resources
References
- Collin M & Bidley V. (2018) Human dendritic cell subsets: an update. Immunology 154(1): 3–20.
- Kozlova AA et al. (2020) Changes in Autofluorescence Level of Live and Dead Cells for Mouse Cell Lines. J Fluoresc 30(6): 1483–9.
- Perfetto SP et al. (2006) Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J Immunol Methods 313(1-2): 199–208.
- Mjösberg JM et al. (2011) Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol 12(11): 1055–62.
- Halim TYF & Takei F. (2014) Isolation and characterization of mouse innate lymphoid cells. Curr Protoc Immunol 106: 3.25.1–13.
- Micossé C et al. (2019) Human “T H 9” cells are a subpopulation of PPAR-γ + T H 2 cells. Sci Immunol 4(31): eaat5943.
- Reighard SD et al. (2020) Therapeutic Targeting of Follicular T Cells with Chimeric Antigen Receptor-Expressing Natural Killer Cells. Cell Rep Med 1(1): 100003.
- Duurland CL et al. (2017) CD161 + Tconv and CD161 + Treg Share a Transcriptional and Functional Phenotype despite Limited Overlap in TCRβ Repertoire. Front Immunol 8: 103.
- Cruz Tleugabulova M et al. The Protein Phosphatase Shp1 Regulates Invariant NKT Cell Effector Differentiation Independently of TCR and Slam Signaling. J Immunol 202(8): 2276-86.
- Nowyhed HN et al. (2017) ATP Binding Cassette Transporter ABCA7 Regulates NKT Cell Development and Function by Controlling CD1d Expression and Lipid Raft Content. Sci Rep 7: 40273.
- Antignano et al. (2014) Methyltransferase G9A regulates T cell differentiation during murine intestinal inflammation. J Clin Invest 124(5): 1945-55.
- Zhao Y et al. (2016) Phenotype, development, and biological function of myeloid derived suppressor cells. Oncoimmunology 5(2): e1004983.
- Nestorowa S et al. (2016) A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128(8): e20-31.
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration