Transitioning Pluripotent Stem Cells from Feeder-Free Media to mTeSR™ Plus
Transitioning human embryonic stem (ES) and induced pluripotent stem (iPS) cells from feeder-free media to mTeSR™ Plus, a serum-free, stabilized cell culture medium
The following protocol is for transitioning human embryonic stem (ES) and induced pluripotent stem (iPS) cells from feeder-free media (e.g. mTeSR™1) to mTeSR™ Plus, a serum-free, stabilized cell culture medium. mTeSR™ Plus allows for greater cell expansion rates with daily feeding, while also maintaining cell quality during restricted feeding schedules. For complete instructions, refer to the Technical Manual: Maintenance of Human Pluripotent Stem Cells in mTeSR™ Plus or contact us to request a copy.
Cells maintained as monolayer cultures in TeSR™ medium (e.g. eTeSR™) can transition to aggregate cultures in mTeSR™ Plus. The protocol can be found in this Tech Tip: Aggregate Passaging for ES and iPS Cell Maintenance, which describes methods using either Gentle Cell Dissociation Reagent (GCDR; Catalog #100-0485) or ReLeSR™ (Catalog #100-0484).
Materials
- Cell Culture Matrix (e.g. Vitronectin XF™, Catalog #07180, or Corning® Matrigel®)
- 6-well Tissue Culture Plate. The type of plate required will depend on your desired matrix:
- With Vitronectin XF™, use non-tissue culture-treated cultureware (e.g. Non Tissue Culture-Treated 6-Well Plates, Catalog #27147)
- With Corning® Matrigel®, use tissue culture-treated cultureware (e.g. Falcon® 6-Well Flat-Bottom Plate, Tissue Culture-Treated, Catalog #38016)
- Dissociation Reagent (e.g. ReLeSR™, Catalog #05872, or Gentle Cell Dissociation Reagent Catalog #07174)
- mTeSR™ Plus (Catalog #05825)
- mTeSR™1 (Catalog #85850)
Protocol
-
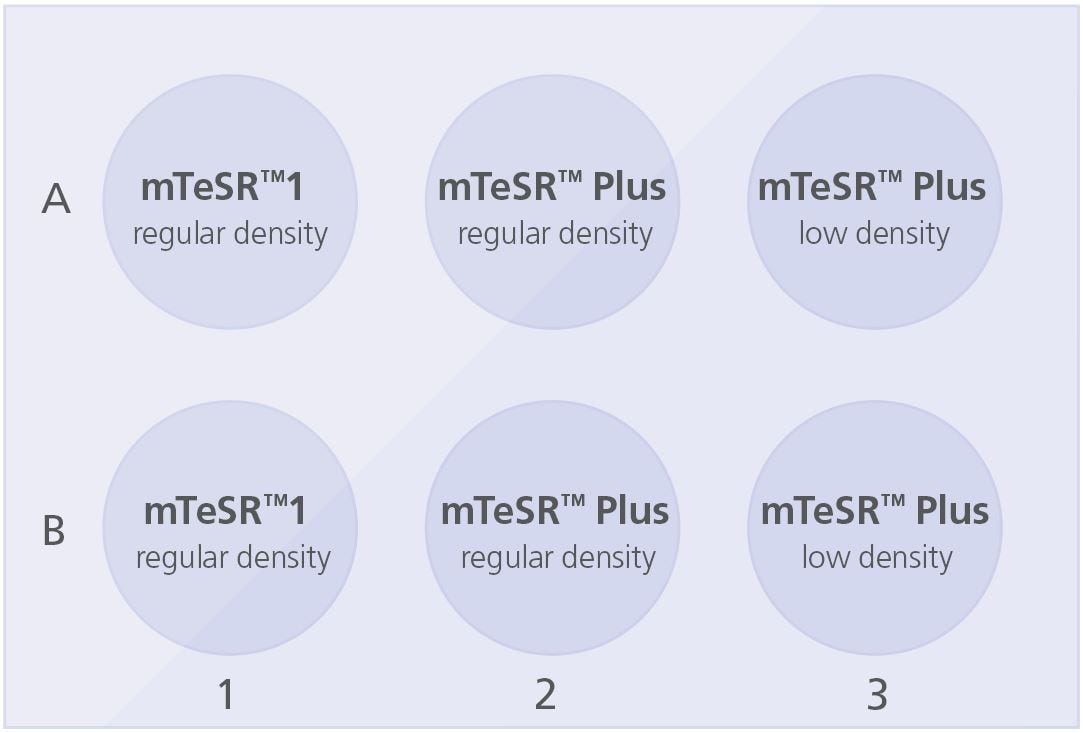
Coat a 6-well tissue culture plate with Corning® Matrigel® (section 4.2.1) or Vitronectin XF™ (section 4.2.2). Add 2 mL medium/well to the coated plate as indicated below:
Note: It is important to maintain mTeSR™1 conditions in parallel until mTeSR™ Plus cultures are established. - Generate a cell aggregate solution using either ReLeSR™ (section 5.1) or Gentle Cell Dissociation Reagent (section 5.2).
-
Plate the cell aggregate mixture as follows:
- For wells in column 1: Use current mTeSR™1 plating density/split ratio
- For wells in column 2: Use current mTeSR™1 plating density/split ratio
- For wells in column 3: Decrease current plating density by ~25%
- Place the plate in a 37°C incubator. Move the plate in several quick, short, back-and-forth and side-to side motions to distribute the cell aggregates. Do not disturb the plate for 24 hours.
-
Perform medium changes as indicated below. Visually assess cultures every 1 - 2 days to monitor growth.
- mTeSR™1: Perform daily medium changes.
- mTeSR™ Plus: Medium can be changed daily or every other day. To skip two consecutive days of feeding, add twice the volume of medium.
- Based on the visual assessment (section 3.2), determine when the cultures are ready to passage. The appropriate passaging day for the mTeSR™ Plus cultures may be earlier than with mTeSR™1 cultures.
-
At the time of passaging, select the mTeSR™ Plus well that is ready to be split; maintain the split ratio of the selected well. If overgrowth is observed, decrease plating density by 20%.
Note: The dissociation incubation times may need to be adjusted; dissociation with non-enzymatic passaging reagents should be monitored under the microscope until the optimal time is determined.
Related Resources
Quality Control for Pluripotent Stem Cells
Get to know the key quality attributes of hPSC cultures, including techniques for maintaining and assessing genomic integrity, pluripotency, and morphology.
Learn More >Controlling the hPSC Culture System
Reducing stressors and undefined components in culture is critical for the maintenance of high-quality hPSCs. Explore resources on feeder-free culture and handling techniques.
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration