Hematopoietic Stem and Progenitor Cells (HSPCs): Isolation, Culture, and Assays
This review provides an overview of HSPC research with a focus on methods of isolation, culture, and assays used to detect and enumerate HSPCs
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration
- Document # 29068
- Version 2.0.0
- Nov 2024
Introduction
Most blood cells have a finite lifespan and are replaced throughout life by the proliferation and differentiation of a very small population of pluripotent hematopoietic stem cells (HSCs). HSCs make up a very small population of the hematopoietic system, however, these cells possess the unique ability to self-renew and undergo successive stages of differentiation, ultimately giving rise to mature blood cells (Figure 1). Also making up a part of the hematopoietic system are short-term repopulating progenitor cells, which give rise to lineage-specific cell types. Collectively, these are referred to as hematopoietic stem and progenitor cells (HSPCs).
The bone marrow (BM) is the primary site of hematopoiesis in humans after birth. Certain treatments, such as cytokine administration (notably granulocyte colony-stimulating factor; G-CSF), myelosuppressive drugs used in cancer therapy, and agents disrupting hematopoietic-BM stromal cell interactions, facilitate rapid mobilization of stem and progenitor cells into circulation. Transplantation of BM or mobilized peripheral blood (mPB) from HLA-matched donors is a well-established therapy for leukemia and various hematologic and immune disorders. Moreover, umbilical cord blood (CB) obtained at birth and cryopreserved provides an alternative source of HSPCs, particularly for patients without suitable BM or mPB donors. Read more about the different sources of HSPCs here.
Recent technological advances in maintaining or amplifying HSPC populations in culture, alongside CRISPR/Cas9 gene editing techniques, have catalyzed the emergence of innovative cell and gene therapy approaches to treat genetic blood disorders like sickle cell disease and thalassemia. The development of cell and gene therapy applications depends on the progress of fundamental research into HSPC biology, predominantly explored through murine models, and the refinement of methodologies for HSPC identification, isolation, and functional characterization from diverse tissue sources, such as bone marrow, blood, and other tissues.
This review aims to provide a comprehensive overview of transplantation and culture assays devised to evaluate the hematopoietic potential of HSPCs. It further delves into phenotypic markers and methodologies for isolating mouse and human HSPCs. Additionally, the review explores culture systems tailored to expand limited HSPC pools in clinical grafts, promote HSPC proliferation and lineage-specific differentiation toward mature blood cell lineages, and generate HSPCs and blood cells from pluripotent stem cells (PSCs).
In Vivo Assays for Hematopoietic Stem and Progenitor Cells
An important characteristic of an HSC lies in its ability to restore blood cell production after transplantation. Experimental designs involving the transplantation of BM cells or purified HSPC subsets in mice have proven invaluable for understanding the biology of both mouse and human stem cells.
Transplantation Studies Using Mouse Cells
To assess the hematopoietic potential of mouse HSCs, BM cells are typically injected into mice whose hematopoiesis has been suppressed, commonly through irradiation. The subsequent repopulation of recipient mice with cells derived from the donor is monitored periodically in blood samples to examine repopulation kinetics. Furthermore, after a minimum of four months, donor-derived cells are evaluated in various tissues including BM, spleen, blood, and thymus to ascertain long-term engraftment (Szilvassy et al., 2002).
The transplantation assay format varies in the genetic backgrounds of donor and host mouse strains, the methodology employed to ablate or suppress hematopoiesis in the recipient pre-transplantation, the detection techniques utilized to identify the progeny of donor-derived stem cells, and the defined endpoints and criteria for determining "successful" engraftment.
In one type of assay, recipient mice undergo total body irradiation to eliminate HSPCs. Subsequently, these mice receive intravenous injections of BM cells or purified HSPCs from a donor mouse, along with "competitor" cells from the same mouse strain as the recipient mice (Purton et al., 2000). These competitor cells serve multiple purposes: providing short-term radioprotection, ensuring recipient survival, and exerting selective pressure to identify stem cells with robust competitive repopulating potential. Alternatively, other assays utilize recipient mice with defective endogenous hematopoiesis due to mutations in the Kit gene. In these models, transplantation with donor cells from wild-type mice is feasible without the need for co-transplanted radioprotective cells (Miller & Eaves, 1997).
The prevalent method for tracing the progeny of transplanted HSCs involves using donor and recipient mouse strains expressing different isoforms of the pan-leukocyte antigen CD45 (CD45.1 or CD45.2). This distinction enables the differentiation between donor and recipient cells via flow cytometry (Spangrude et al., 1988). While this technique effectively identifies donor-derived lymphocytes, granulocytes, monocytes, and HSPCs, it cannot detect donor-derived erythrocytes and platelets, as these cells lack CD45 expression. However, by employing transgenic donor cells expressing a readily detectable reporter molecule, such as green fluorescent protein (GFP), in all blood cell lineages, including the erythroid lineage, the engraftment of transplanted cells can be quantified across all cellular lineages (Boyer et al., 2019).
In this experimental setup, different cohorts of mice receive varying quantities of donor cells. After several months, the extent of hematopoietic reconstitution in each group of mice is evaluated. Utilizing Poisson statistics, researchers can then determine the frequency of repopulating cells within the transplanted cell population (Szilvassy et al., 1990). This methodology, complemented by single-cell transplantations wherein purified HSPCs are individually injected into groups of mice, facilitates a detailed quantitative analysis of HSC frequencies across diverse purified HSPC subsets. Moreover, it enables the investigation of engraftment dynamics and differentiation potential at the individual HSPC level. To investigate the in vivo self-renewal capacity of mouse HSCs, researchers can also perform serial transplantations, wherein bone marrow cells from primary recipients are transplanted into secondary, tertiary, and even quaternary recipients.
Recent advancements have revolutionized the study of HSCs by enabling the labeling of millions of individual cells within a heterogeneous population with unique genetic markers (“barcodes”). These markers, integrated into the genome, are inherited by subsequent generations of daughter cells. Upon transplantation of numerous tagged HSCs into recipient mice, the engraftment kinetics, size, and lineage distribution of each labeled HSC can be precisely quantified. This is achieved by identifying the specific barcode within blood cells using high-throughput parallel sequencing methods (Naik et al., 2014). This approach facilitates a comprehensive clonal analysis of multiple HSCs, overcoming the inherent limitations of traditional single-cell transplantation experiments.
Transplantation Studies Using Human Cells
The hematopoietic potential of human HSPCs can be investigated through transplantation into genetically immune-deficient mice, followed by the assessment of human blood cells in the blood, BM, or other organs of these mice several weeks to months later.
Traditionally, these assays were conducted using the severe combined immunodeficiency (SCID) and non-obese diabetic (NOD)/SCID mouse strains. However, these approaches required large cell numbers to overcome immune rejection by residual host macrophages and natural killer (NK) cells. Moreover, these assays exhibited a bias towards B-lymphocyte development, while impeding sustained long-term production of myeloid cells, platelets, and erythrocytes. Consequently, studying the kinetics of human cell engraftment or distinguishing between short-term and long-term reconstitution mediated by distinct HSC subsets was not possible.
The development of other immunodeficient mouse strains has alleviated many of these challenges. Specifically, β2-microglobulin-deficient and interleukin-2 receptor (IL-2R) γ-deficient NOD/SCID mice demonstrate robust engraftment levels for over 20 weeks post-transplantation (Cheung et al., 2013; Ito et al., 2002; Kollet et al., 2000; Shultz et al., 2005). IL-2Rγ-deficient mice, coupled with functional impairment of endogenous HSCs due to loss-of-function mutations in the Kit gene, exhibit even greater permissiveness for human HSC engraftment and prevent the need for pre-transplant conditioning via irradiation (Cosgun et al., 2014).
These enhanced xenotransplantation assays have facilitated a more comprehensive characterization of human HSCs and have been instrumental in studies identifying human HSCs at single-cell resolution (Notta et al., 2011). For further insights into the historical context of xenogeneic mouse transplantation assays and the utilization of other mouse strains, please refer to a recent review by Mian et al. (2021).
In Vitro Assays for Hematopoietic Progenitor Cells
While transplantation assays are essential for evaluating the engraftment potential and hematopoietic reconstitution ability of HSPCs in vivo, culture assays have also emerged as valuable tools for assessing the proliferative and differentiation capacities of these cells. These assays are capable of detecting hematopoietic cells at various stages of differentiation, ranging from multipotent progenitors to lineage-restricted progenitor cells. In the subsequent sections, we will delve into the principles and applications of two of the best-characterized culture assays, the colony-forming unit (CFU) assay and the long-term culture-initiating (LTC-IC) assay.

Figure 1. Hematopoietic Hierarchy, Assays and Markers in Mice and Humans
Schematic representation of the hematopoietic hierarchy from HSCs, via intermediate progenitor stages, to mature blood cells. Assays to identify HSCs and progenitors are shown at the top. The most definitive phenotypical markers used to identify the various types of mouse and human hematopoietic cells are shown at the bottom. Additional markers can be used to further distinguish between subsets. Refer to the text for further details. It should be noted that this model is continuously being revised and updated and several other ways of presenting the hematopoietic hierarchy and differentiation trajectories have been proposed (Laurenti & Gottgens, 2018). For example, HSCs are more heterogeneous with respect to self-renewal and differentiation abilities, and lineage commitment and differentiation are more continuous than suggested by surface marker expression profiles and functional criteria alone. The model also doesn't show the vast differences in the numbers of individual types of blood cells generated, with erythrocytes and neutrophils representing ~97% of the ~300 billion new blood cells (excluding platelets) produced in the human bone marrow every day under steady-state conditions. (Sender & Milo, 2021).
LT-HSC: Long-Term Hematopoietic Stem Cell; ST-HSC: Short-Term Hematopoietic Stem Cell; MPP: Multipotential Progenitor; CMP: Common Myeloid Progenitor; CLP: Common Lymphoid Progenitor; CFU-GEMM: Colony-Forming Unit – Granulocyte/Erythrocyte/Macrophage/Megakaryocyte; BFU-E: Burst-Forming Unit – Erythroid; CFU-E: Colony-Forming Unit – Erythroid; CFU-Mk: Colony-Forming Unit – Megakaryocyte; CFU-GM: Colony-Forming Unit – Granulocyte/Macrophage; CFU-G: Colony-Forming Unit –Granulocyte; CFU-M: Colony-Forming Unit – Macrophage
Colony-Forming Unit Assays
Since its inception over five decades ago (Bradley & Metcalf, 1966), the colony-forming unit (CFU) assay, also called colony-forming cell (CFC) assay, has emerged as the gold standard in vitro functional assay for investigating hematopoietic progenitor cells. In the CFU assay, cells are cultured at low cell density in a semi-solid medium, typically methylcellulose-based (e.g. MethoCult™), and supplemented with appropriate cytokines. These conditions support the proliferation and differentiation of individual progenitor cells, or CFUs, which give rise to discrete colonies in usually one to two weeks of culture. Distinct colonies arising from various types of progenitor cells are identified and quantified based on the number and types of mature cells they contain, utilizing morphological and phenotypic criteria. The CFU assay is primarily employed to detect multipotential and lineage-restricted progenitors of the erythroid, granulocytic, and macrophage lineages. Moreover, megakaryocyte and B-lymphoid progenitors can also be identified by applying selective culture conditions tailored to these specific cell types.
Widely utilized across research and clinical settings, the CFU assay is an important tool for examining the impacts of stimulatory and inhibitory agents on HSPC proliferation and differentiation, including potential toxicities of new drugs. Learn more in our whitepaper, The CFU Assay in Preclinical Toxicity Testing. Additionally, it facilitates the evaluation of in vitro manipulations, such as cell processing, cryopreservation, genetic modification, and ex vivo expansion culture, thereby ensuring the quality of cellular products utilized in hematopoietic cell transplantation and other therapeutic interventions.
Multiple studies have linked CFU counts to key transplantation outcomes, such as engraftment time and overall survival post-transplantation (Hogge et al., 2000; Iori et al., 2004; Page et al., 2011; Prasad et al., 2008; Yang et al., 2005; Yoo et al., 2007). Consequently, this assay is valuable for predicting graft quality, notably aiding in selecting cord blood units with viable progenitor cells before transplantation. Automated imaging platforms, such as STEMvision™, streamline colony counting, enhancing reproducibility and standardization (Velier et al., 2019).
Training: Introduction to STEMvision™ for Automated CFU Scoring
Learn how to standardize and automate the scoring of the CFU assay using STEMvision™ in this self-paced digital course. You’ll gain access to lectures, step-by-step protocol videos, and a library of curated resources.
Start Now >
Long-Term Culture Assay for Primitive Hematopoietic Progenitor Cells
The long-term culture (LTC) assay identifies more primitive progenitor cells than the CFU assay. The initial versions of the LTC assay developed in the late 1970s were designed to detect primitive progenitors of the myeloid (i.e. granulocyte, macrophage, erythroid, and megakaryocyte) lineages (Dexter et al., 1977; Gartner & Kaplan, 1980). Later advancements also facilitated the growth and identification of B lymphoid and NK cell progenitors (Miller et al., 1992; Whitlock & Witte, 1982).
In executing the LTC assay, hematopoietic cells are cultured atop an adherent monolayer derived from primary bone marrow or immortalized stromal cell lines. Using specialized culture media, such as MyeloCult™, this system fosters the survival, self-renewal, proliferation, and differentiation of primitive hematopoietic cells, including long-term repopulating HSCs, over extended periods spanning many weeks (Cho & Muller-Sieburg, 2000; Miller & Eaves, 2002).
The cells identified in LTC assays are termed long-term culture-initiating cells (LTC-ICs). These cells are typically distinguished by their ability to generate CFUs in the stroma-supported cultures for at least five weeks (four weeks for mouse cells). This duration ensures that any CFUs in the original cell sample undergo terminal differentiation. Consequently, CFUs detected after 4 - 5 weeks are newly generated from more primitive cells. The LTC-IC-derived CFUs are identified by re-plating the contents of individual cultures in CFU assay media (such as MethoCult™) and counting colonies approximately two weeks later (Miller & Eaves, 2002).
For optimal accuracy, LTC-IC assays ideally utilize a limiting-dilution design that enables measurement of the frequency of these progenitor cells. Alternatively, simple formats that measure the CFU output of bulk cultures can also determine the number of LTC-ICs if the CFU yield per LTC-IC is already established from prior studies. In a different iteration, individually purified primitive HSPCs are sorted into microwells containing stromal cells and culture medium. The clonal lineage output is assessed six weeks later (Knapp et al., 2018).
Markers and Methods for HPSC Isolation
In addition to the functional assays described in the previous paragraphs, HSPCs can also be distinguished based on their phenotype. This involves staining cells, typically with fluorescently labeled antibodies targeting cell surface antigens or DNA dyes, and subsequently analyzing through flow cytometry or microscopy. This method has greatly enhanced our understanding of key markers distinguishing HSCs, progenitors, and mature blood cells. Such insights have been essential for devising strategies to isolate HSCs from bone marrow (BM), blood, and other heterogeneous cell populations.
This section will delve into the most commonly utilized markers for identifying and purifying HSCs in mice and humans. Additionally, a summary detailing additional markers utilized to differentiate progenitor subsets, including common myeloid progenitors, common lymphoid progenitors, granulocyte and macrophage progenitors, and megakaryocyte and erythroid progenitors, was previously provided in Figure 1.
Mouse Cells
The initial step in identifying and isolating mouse HSPCs in BM, spleen, fetal liver, or other tissues usually involves the elimination of mature cells that express “lineage" (Lin) antigens specific to terminally differentiated blood cells. Lin antigens, such as CD3 for T cells, B220 for B cells and NK cells, Ly6G/Gr-1 for granulocytes, CD11b/Mac-1 for monocytes and macrophages, and TER-119 for erythroid cells, are absent or expressed weakly on HSPCs.
Following the elimination of lineage-positive (Lin+) cells through immunomagnetic isolation methods like EasySep™, HSPCs can be further characterized using combinations of cell surface markers, notably KIT (i.e. the receptor for stem cell factor [SCF], also known as CD117) and SCA1 (Muller-Sieburg et al., 1986; Okada et al., 1992; Osawa et al., 1996; Spangrude et al., 1988; Uchida et al., 2003).
Lin-SCA1+KIT+ (LSK) cells account for less than 0.1% of nucleated BM cells but contain most repopulating HSCs. However, SCA1 is only useful in some mouse strains (e.g. C57Bl/6), but not in BALB/c and other mouse strains which exhibit low SCA1 expression on HSCs (Spangrude & Brooks, 1992, 1993). Alternatively, markers and isolation strategies based on a Lin-CD48-CD150+ phenotype (the SLAM phenotype) are effective across most mouse strains (Kiel et al., 2005).
LSK and SLAM cells represent heterogeneous populations, with HSCs accounting for at most 10 - 20% of all cells. Further enrichment of HSCs, achieving frequencies as high as 50%, has been accomplished by selecting LSK or SLAM cells expressing elevated levels of CD201 (endothelial protein C receptor; EPCR) and low or undetectable levels of CD34, CD135 (FLT3), and CD49b. These enriched HSC populations retain low levels of DNA dyes like Rhodamine-123 (Rho123) and Hoechst 33342 due to the high expression of multidrug transporter proteins MDR1 and ABCG2, respectively (Balazs et al., 2006; Benveniste et al., 2010; Kent et al., 2009; Kiel et al., 2005; Osawa et al., 1996; Uchida et al., 2003; Uchida & Weissman, 1992).
Human HSPCs
The cell surface protein CD34 is the most important marker of primitive human hematopoietic cells. CD34 is expressed in 1 - 5% of nucleated human BM cells, approximately 1% of CB cells, and less than 0.1% of normal peripheral blood (PB) cells. Most human HSCs are CD34+, as demonstrated by xenotransplantation assays described above and clinical transplants performed with purified CD34⁺ cells from BM and mPB (Civin et al., 1996; Larochelle et al., 1996; Vogel et al., 2000) The CD34⁺ population also includes most LTC-ICs, CFU-GEMM, BFU-E, and CFU-GM. As cells differentiate, CD34 expression decreases, and the majority of late-stage progenitors (e.g. CFU-E) and mature blood cells become CD34- (Civin et al., 1996; Strauss et al., 1986).
CD34+ cells are enriched 50 to 200-fold by depleting Lin+ cells using methods such as EasySep™, RosetteSep™, or fluorescence-activated cell sorting (FACS). CD34 is expressed on approximately 40 - 90% of Lin- BM cells depending on the individual sample and experimental conditions, and can be further purified using CD34+ isolation methods. HSCs and primitive progenitors that are detectable in xenotransplantation and LTC-IC assays account for only 0.1 - 1% of CD34+ blood or BM cells, while progenitor cells that are detectable in CFU assays comprise about 10 - 30% of CD34+ cells. Therefore, CD34 expression alone is not sufficient to measure HSCs, and additional markers are required to identify and isolate the most primitive hematopoietic cells.
The most common markers to subfractionale CD34+ cells include CD38 and CD45RA, which are absent or only weakly expressed on primitive cells, and CD90 (THY1), which is expressed at higher levels on primitive cells than on differentiated cells (Craig et al., 1993; Lansdorp et al., 1990; Terstappen et al., 1991). As few as 10 Lin-CD34+CD38-CD45RA-CD90+ CB cells can engraft the BM of immunodeficient mice and generate human lymphoid and myeloid cells for at least 12 weeks after transplantation. These results demonstrate that Lin-CD34+CD38-CD45RA-CD90+ cells are highly enriched for long-term repopulating cells. Some CD90- cells can also engraft but at a lower level, and may represent a less primitive HSC subset than their CD90⁺ counterparts (Majeti et al., 2007).
A subset of Lin-CD34+CD38-CD45RA- also express the adhesion molecule CD49f and weakly retain the DNA dye, Rhodamine (Rho)-123 (Notta et al., 2011). In single-cell transplantation studies, close to 30% of the Lin-CD34+CD38-CD45RA-CD90+CD49f+Rho- low exhibited long-term multi-lineage repopulating ability (Notta et al., 2011). A similarly high HSC frequency of 1 out of 3 cells has been observed in limiting dilution transplantation experiments with purified CD34+CD38-CD45RA-EPCR+ cells. These cells are heterogeneous for CD90 expression, but nearly all are CD49f+ (Anjos-Afonso et al., 2022).
Human HSPCs can also be identified by measuring the activity of the intracellular enzyme aldehyde hydrogenase (ALDH), specifically the ALDH1a1 isozyme, which is involved in retinoid signaling and detoxification of aldehydes (Wang et al., 2022). ALDH converts a non-charged fluorescent substrate (ALDEFLUOR™), which passively crosses cell membranes, into a negatively charged polar fluorescent product, which cannot cross cell membranes and accumulates inside viable, intact ALDH-expressing cells. As a result, cells with high ALDH activity can be detected and isolated by flow cytometry based on their bright fluorescence (Storms et al., 1999). Both long- and short-term repopulating HSCs and most progenitor cells, with the possible exception of lymphoid progenitors, express high levels of ALDH (Hess et al., 2004; Storms et al., 2005). Flow cytometric methods to measure the frequency of CD34brightALDH-bright cells have been developed as a potency assay to measure the quality of CB units intended for transplantation (Shoulars et al., 2016).
The aforementioned markers and isolation strategies are vital for the isolation of HSPCs from diverse cell populations such as BM, mPB, or CB. Apart from CD38, which is primarily useful for distinguishing primitive and differentiated cells in noncultured cells, these markers play a crucial role in identifying HSPCs in cell culture experiments. In the subsequent section, we will explore various examples of HSPC culture methods. These encompass techniques aimed at (i) augmenting the limited numbers of HSPCs in grafts; (ii) activating HSPCs for gene editing purposes; (iii) generating mature blood cells in lineage-specific CD34+ cell cultures; and (iv) inducing hemogenic specification and differentiation of pluripotent stem cells to produce lineage-specific progenitor cells and mature blood cells.
Hematopoietic Stem and Progenitor Cell Expansion Cultures
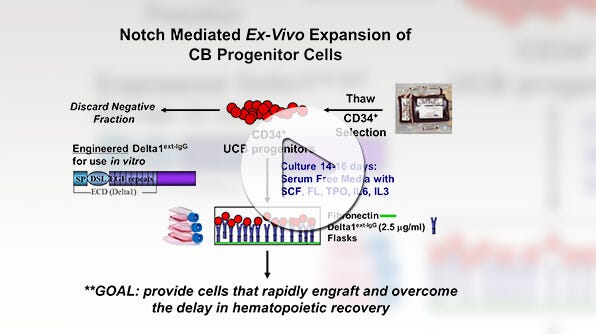
Umbilical cord blood (CB) serves as an alternative source of HSCs for transplantation in patients lacking a suitable BM or mPB donor. However, most CB units lack sufficient numbers of primitive cells to facilitate successful engraftment in adult recipients. To overcome this challenge, extensive research has focused on developing culture methods capable of augmenting the numbers and engraftment potential of repopulating cells, aiming to enhance the clinical applicability of CB for cellular therapy.
One approach to increase HSPC numbers involves culturing CD34+ cells with combinations of cytokines, such as IL-3, IL-6, SCF, Flt3L, TPO, GM-CSF, and G-CSF. This method has demonstrated a remarkable capability to expand CD34+ cell numbers by 100 to 1000-fold within 7 - 20 days of culture. However, typical culture conditions tend to induce differentiation in most HSPCs, resulting in the loss of primitive HSPCs and their replacement with more differentiated cells, which may outcompete any remaining HSPCs by the end of the culture period.
To enhance the frequency and yields of primitive HSPCs post-culture, several strategies have been devised. These include culturing CD34+ cells or highly purified primitive HSPCs at low cell densities, with frequent medium changes or in fed-batch cultures, to mitigate the impact of inhibitory factors secreted by differentiating cells (Csaszar et al., 2012). Other advancements involve the utilization of supportive mesenchymal stromal cells (de Lima et al., 2012), immobilized Notch-Ligand (Delaney et al., 2010), or the incorporation of specific soluble small molecule HSC agonists into the culture medium. Noteworthy small molecule agonists whose effectiveness has been validated include nicotinamide, StemRegenin 1 (SR1), and UM171 (or its analog, UM729) (Boitano et al., 2010; Fares et al., 2014; Peled et al., 2012; Subramaniam et al., 2020). Supplementing CD34+ cultures with small molecule agonists has been shown to elevate HSC numbers by approximately 10-fold, as determined by transplantation assays in immunodeficient mice, compared to cultures with cytokines alone.
Some of the enhanced HSPC expansion techniques have been successfully implemented in clinical trials to improve the engraftment of transplanted CB CD34+ cells in patients with hematologic malignancies. Specifically, HSPCs expanded with cytokines, along with either nicotinamide or UM171, supported earlier neutrophil and platelet recovery along with improved long-term multi-lineage engraftment compared to single unmanipulated CB transplantation (Cohen et al., 2020; Horwitz et al., 2021). The sustained engraftment of culture-expanded HSCs relied on the co-infusion of donor T cells, which likely prevent the elimination of cultured HSCs by residual patient T cells or, in the case of double CB transplantation, by donor T-cells from a second unmanipulated graft (Cohen et al., 2020; Horwitz et al., 2014; Wagner et al., 2014). These studies offer compelling evidence that ex vivo HSPC expansion can be advantageous for clinical transplantation.
Gene Editing of HSPCs
Improvements in HSPC culture methods, as outlined in the preceding paragraph, have not only advanced transplantation techniques but also facilitated the development of other applications for cellular therapy involving HSPCs. A notable example is the genetic modification of BM and mPB-derived CD34+ cells for the treatment of inherited blood and immune disorders, such as beta-thalassemia, sickle cell disease, immunodeficiencies, and lysosomal storage disorders, which can be managed through HSC transplantation.
Gene editing strategies, employing lentiviral vector transduction or the more recent CRISPR-Cas-mediated gene editing technology, necessitate the activation of quiescent HSPCs under conditions that maintain HSC numbers and functions. The culture media, cytokines, and HSC agonists utilized in gene editing workflows are similar to those developed for the expansion of CB CD34+ cells. However, shorter culture times of 2 - 4 days are preferred to achieve high gene editing efficiencies in BM and CB CD34+ cells without necessarily augmenting HSC numbers beyond input levels. This duration is notably shorter than the 7 - 21 day cultures required for CB CD34+ cells to attain clinically relevant HSC numbers (Gentner et al., 2021; Kohn et al., 2020; Lattanzi et al., 2021; Rai et al., 2023).
Webinar: Optimizing CD34+ Cell Genome Editing for Efficiency and HSPC Maintenance
Learn tips and tricks for effective genome editing of CD34⁺ cells, methods to evaluate genome editing efficiency, as well as optimal pre- and post- editing culture conditions to maintain HSPC function and long-term editing effects.
Generation of Mature Blood Cells in Culture
In addition to the expansion of HSCs themselves, researchers have developed culture methods to promote the lineage-specific differentiation of HSPCs, aiming to generate large numbers of mature blood cells, including monocytes/macrophages, lymphoid cells (T, B, and NK cells), erythroid cells, megakaryocytes, and platelets.
These HSPC differentiation cultures serve various purposes, including evaluating lineage-specific toxicity of novel drug candidates, assessing the capability of culture-expanded or gene-edited HSPCs to generate functional blood cells, and generating permissive target cells for reprogramming hematopoietic cells into induced pluripotent stem cells (iPSCs). Additionally, larger-scale differentiation cultures have been established to yield substantial quantities of blood cells, such as red blood cells (RBCs) and platelets, with the potential to replace donated blood products in transfusions
In the subsequent paragraphs, we will explore two methods for promoting erythroid and megakaryocyte differentiation of HSPCs.
Culture of Red Blood Cells
HSPCs can be stimulated to proliferate and differentiate into erythroid cells through culture in serum-free media supplemented with SCF, IL-3, and EPO, in conjunction with a glucocorticoid receptor (GR) agonist like hydrocortisone (Leberbauer et al., 2005). Erythroid progenitors exhibit a remarkably high proliferative capacity, yielding large quantities of erythroblasts over 1 - 3 weeks of culture. The most substantial cell yields, typically ranging from 105 to 106 cells per CD34+ cell, are often obtained in cultures of purified CB CD34+ cells. However, bone marrow and non-mobilized peripheral blood mononuclear cells (PBMCs) also yield favorable results.
In typical expansion cultures, most erythroid cells comprise CD71+GpA+ erythroblasts with minimal hemoglobin expression. Transitioning to a culture medium containing only EPO, excluding IL-3, SCF, or GR agonist, promotes further differentiation, as characterized by reduced cell size, down-regulation of CD71, hemoglobinization, and enucleation (Leberbauer et al., 2005). The maturation efficiency of culture-expanded erythroblasts into reticulocytes and erythrocytes can be enhanced by co-culturing with immortalized or bone marrow-derived stromal cells, or by incorporating various supplements, such as serum or plasma, high transferrin concentrations, or heparin (Gallego-Murillo et al., 2022; Giarratana et al., 2005; Leberbauer et al., 2005; Miharada et al., 2006). Cultured RBCs exhibit similar oxygen-binding properties to native RBCs and demonstrate survival in immunodeficient mice and human recipients (Giarratana et al., 2005). Several research groups have devised scaled-up versions of erythroid cultures in bioreactors to produce RBCs for clinical testing, although further refinements are essential to manufacture RBCs in sufficient quantities for routine use in transfusion medicine (Pellegrin et al., 2021).
Culture of Megakaryocytes and Platelets
Culture conditions have also been developed to promote the differentiation of HSPCs into megakaryocytes and platelets. Thrombopoietin (TPO) is the primary regulator of megakaryocytopoiesis and can facilitate the differentiation of HSPCs into megakaryocytes (MKs) and platelets in cultures of CB, BM, or mPB CD34+ cells. However, employing a combination of cytokines, including SCF, Flt3L, IL-1ß, IL-3, IL-6, IL-9, IL-11, and GM-CSF, alongside TPO, yields better results, typically producing around 100 MKs per original CD34+ cell. When individual cytokines are meticulously titrated to prevent the outgrowth of granulocyte and macrophage progenitors, approximately 90% of cultured cells express MK antigens, such as CD41 and CD42 (L. Boyer et al., 2008; Cortin et al., 2005). Similar to MKs in BM, culture-derived MKs are large and polyploid and can generate platelet-like particles through proplatelet formation.
Scaling up MK cultures to achieve therapeutically useful cell numbers is a challenge even bigger than for erythroid cultures. This is primarily because MK progenitors are less abundant and generate fewer cells than erythroid progenitors, with cell yields exhibiting significant variability among CD34+ cells from individual CB, BM, or mPB donors.
To tackle these challenges, researchers are increasingly exploring novel sources of HSPCs and blood cells, particularly embryonic stem cells (ESCs) and iPSCs, collectively termed pluripotent stem cells (PSCs). The following section will describe methods for inducing the differentiation of PSCs into hematopoietic cells.
Several differentiation protocols have been developed to generate hematopoietic cells from PSCs. In one approach, PSCs are plated as clumps or single cells on a layer of solubilized basement membrane matrix components (e.g. Matrigel®) to form a two-dimensional cell layer. In another method, PSCs are aggregated to form 3-dimensional embryoid bodies. In both methods, PSCs undergo initial differentiation into mesodermal cells during 3 - 4 days of culture with cytokines, such as vascular endothelial growth factor (VEGF), bone morphogenetic protein 4 (BMP4), or basic fibroblast growth factor (bFGF).Subsequently, hematopoietic growth factors such as SCF, TPO, IL-3, and IL-6 are introduced to drive the differentiation of mesodermal cells into hematopoietic progenitors. This phase of culture typically spans 7 - 9 days, following which cells are harvested. The cells are then assessed by flow cytometry to identify cells expressing hematopoietic markers, notably CD34, CD45, and CD43, and their ability to generate myeloid, erythroid, and multi-lineage colonies is evaluated in the CFU assays.
Hematopoietic differentiation methods for PSCs can be used for disease modeling and therapy development for leukemias and other hematologic disorders, utilizing PSCs reprogrammed directly from patient cells (Imeri et al., 2023; Kotini et al., 2023). PSC-derived hematopoietic cells can also be cultured under lineage-specific conditions to induce further differentiation, yielding relatively pure populations of mature blood cells, including erythroid cells, megakaryocytes, monocytes/macrophages, and lymphocytes (Bernecker et al., 2019; Feng et al., 2014; Martins et al., 2023; Mesquitta et al., 2019; Shukla et al., 2017). Scaled-up versions of these lineage-specific cultures could potentially be used to produce large quantities of cells for transfusion and immunotherapy applications, provided that the yields and quality of these cells match those of primary hematopoietic cells. Ongoing research efforts are focused on enhancing the efficiency and fidelity of PSC-based hematopoietic differentiation protocols to achieve this goal.
Considerable research efforts have also been directed at method development to generate PSC-derived cells that can reconstitute hematopoiesis after transplantation. Most PSC-derived hematopoietic cells do either not engraft or only show transient myeloid-restricted engraftment after transplantation into immunodeficient mice. The formation of functional HSCs with long-term multilineage stem cell potential has until recently only been achieved with hematopoietic cells derived from genetically modified PSC lines that overexpress transcription factors regulating HSC self-renewal, such as HoxB4, RunX1, and SPi1, or via the formation of PSC-derived germ cell tumors, i.e. teratomas, in mice (Kyba et al., 2002; Sugimura et al., 2017; Suzuki et al., 2013). For practical and safety reasons, methods involving transgene overexpression or tumor formation are unsuitable for the development of clinically useful protocols. However, a recent study has demonstrated that HSCs with multilineage engraftment potential in immunodeficient mice can also be derived from PSCs without the need for transgene overexpression or teratoma formation. The culture conditions employed in this study promote the formation of complex structures akin to the hematopoietic environment supporting HSC formation in the human embryo (Piau et al., 2023). These promising findings underscore the capability of PSCs to differentiate into functional HSCs when exposed to appropriate microenvironmental signals.
Due to the complexity of the culture system, it may be difficult to fully harness the potential of PSCs as an effective cell source for HSC-based cell therapies. Further scientific advancements will be needed to enhance our understanding of the molecular mechanisms governing HSC development, refine and scale up PSC differentiation protocols, and realize the clinical potential of PSC-derived HSCs.
Summary And Conclusions
HSCs are multipotent cells that support the lifelong production of all blood and immune cells. They can be identified by their ability to reconstitute long-term hematopoiesis in transplantation assays, typically performed in mice. Hematopoietic progenitors, which are the intermediate stages between HSCs and blood cells, do not have long-term repopulating ability after transplantation and are measured in culture assays, such as the CFU assay, and LTC-IC assay. Flow cytometry and other antibody-based techniques further aid in the characterization and purification of hematopoietic cells by discerning expression patterns unique to HSCs, progenitors, and mature blood cells.
These assays are instrumental in advancing our comprehension of hematopoiesis and assessing the effects of ex vivo manipulations like purification, cryopreservation, culture, and drug exposure on HSPC numbers, viability, and function. Recent advancements in single-cell resolution techniques to measure chromatin structure, gene and protein expression, and metabolism have greatly enhanced HSPC characterization, offering profound insights into hematopoietic hierarchy dynamics and responses to stimuli.
While donor bone marrow, mobilized blood, and umbilical cord blood are the primary sources of HSPCs for research and clinical purposes, ex vivo expansion culture protocols have been devised to amplify the limited numbers of obtainable HSCs from these sources. The ability to expand HSPCs in culture is crucial for the success of clinical applications, including gene therapy, where sustained HSPC functionality in vitro is vital.
Furthermore, the advancement of technologies enabling the generation of HSPCs from pluripotent stem cells offers an alternative avenue for producing HSCs and blood cells for cellular therapies. Once technical hurdles are surmounted and functional HSCs and blood cells can be manufactured at scale, these innovative methodologies hold immense promise for the development of novel applications. These applications are anticipated to encompass the creation of universal donor cells suitable for transplantation or transfusion, thereby addressing limitations associated with donor availability and compatibility. Additionally, they may facilitate the engineering of immune cells tailored to combat challenging conditions, such as infections and cancers resistant to conventional treatments. By harnessing the potential of tissue-derived and pluripotent stem cell-derived HSPCs, future therapeutic strategies stand to revolutionize the treatment landscape and significantly improve patient outcomes.
Resources for Your Hematopoietic Research
Products for Your Hematopoietic Research
References
- Anjos-Afonso F et al. (2022) Single cell analyses identify a highly regenerative and homogenous human CD34+ hematopoietic stem cell population. Nat Commun 13(1): 1–13.
- Balazs AB et al. (2006) Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood 107(6): 2317–21.
- Benveniste P et al. (2010) Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell 6(1): 48–58.
- Bernecker C et al. (2019) Enhanced ex vivo generation of erythroid cells from human induced pluripotent stem cells in a simplified cell culture system with low cytokine support. Stem Cell Rep 7(23):1540–51.
- Boitano AE et al. (2010) Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 329(5997): 1345–48.
- Boyer L et al. (2008) Increased production of megakaryocytes near purity from cord blood CD34+ cells using a short two-phase culture system. J Immunol Methods 332(1–2): 82–91.
- Boyer SW et al. (2019) Clonal and quantitative in vivo assessment of hematopoietic stem cell differentiation reveals strong erythroid potential of multipotent cells. Stem Cell Rep 12(4): 801–15.
- Bradley TR & Metcalf D (1966) The growth of mouse bone marrow cells in vitro. Aust J Exp Biol Med Sci 44(3): 287–99.
- Cheung AMS et al. (2013) Analysis of the clonal growth and differentiation dynamics of primitive barcoded human cord blood cells in NSG mice. Blood 22(18): 3129–37.
- Cho RH & Muller-Sieburg E (2000) High frequency of long-term culture-initiating cells retain in vivo repopulation and self-renewal capacity. Exp Hematol 28(9): 1080–86.
- Civin CI et al. (1996) Highly purified CD34-positive cells reconstitute hematopoiesis. J Clin Oncol 14(8): 2224–33
- Cohen S et al. (2020) Hematopoietic stem cell transplantation using single UM171-expanded cord blood: a single-arm, phase 1-2 safety and feasibility study. The Lancet Hematology 7(2): e134-45.
- Cortin V et al. (2005) Efficient in vitro megakaryocyte maturation using cytokine cocktails optimized by statistical experimental design. Exp Hematol 33(10): 1182–91.
- Cosgun KN et al. (2014) Kit regulates HSC engraftment across the human-mouse species barrier. Cell Stem Cell 15(2): 227–38.
- Craig W et al. (1993) Expression of Thy-1 on human hematopoietic progenitor cells. J Exp Med 177(5): 1331–42.
- Csaszar E et al. (2012) Rapid expansion of human hematopoietic stem cells by automated control of inhibitory feedback signaling. Cell Stem Cell 10(2): 218–29.
- de Lima M et al. (2012) Cord-blood engraftment with ex vivo mesenchymal-cell coculture. N Engl J Med 367(24): 2305–15.
- Delaney C et al. (2010) Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat Med 16(2): 232–6.
- Dexter TM et al. (1977) Conditions controlling the proliferation of hematopoietic stem cells in vitro. J Cell Physiol 91(3): 335–44.
- Kyba M et al. (2002) HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109(1): 29–37.
- Lansdorp PM et al. (1990) Selective expression of CD45 isoforms on functional subpopulations of CD34+ hematopoietic cells from human bone marrow. J Exp Med 172(1): 363–6.
- Larochelle A et al. (1996) Identification of primitive human hematopoietic cells capable of repopulating NOD/SCID mouse bone marrow: implications for gene therapy. Nat Med 2(12): 1329–37.
- Lattanzi A et al. (2021) Development of β-globin gene correction in human hematopoietic stem cells as a potential durable treatment for sickle cell disease. Sci Transl Med 13(598): eabf2444.
- Laurenti E & Gottgens B (2018) From haematopoietic stem cells to complex differentiation landscapes. Nature 553(7689): 418–26.
- Leberbauer C et al. (2005) Different steroids co-regulate long-term expansion versus terminal differentiation in primary human erythroid progenitors. Blood 105(1): 85–94.
- Majeti R et al. (2007) Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell 1(6): 635–45.
- Martins GLS et al. (2023) Evaluation of 2D and 3D erythroid differentiation protocols using sickle cell disease and healthy donor induced pluripotent stem cells. Cells 12(8): 1121.
- Mesquitta WT et al. (2019) UM171 expands distinct types of myeloid and NK progenitors from human pluripotent stem cells. Sci Rep 9(1): 6622.
- Mian SA et al. (2021) Advances in human immune system mouse models for studying human hematopoiesis and cancer immunotherapy. Front Immunol 11: 619236.
- Miharada K et al. (2006) Efficient enucleation of erythroblasts differentiated in vitro from hematopoietic stem and progenitor cells. Nat Biotechnol 24(10): 1255–6.
- Miller L & Eaves J (1997) Expansion in vitro of adult murine hematopoietic stem cells with transplantable lympho-myeloid reconstituting ability. Proc Natl Acad Sci USA 94(25): 13648–53.
- Miller CL & Eaves CJ (2002) Long-term culture-initiating cell assays for human and murine cells. Methods Mol Med 63: 123–41.
- Miller JS et al. (1992) The generation of human natural killer cells from CD34+/DR- primitive progenitors in long-term bone marrow culture. Blood 80(9): 2182–7.
- Muller-Sieburg CE et al. (1986) Isolation of two early B lymphocyte progenitors from mouse marrow: a committed pre-pre-B cell and a clonogenic Thy-1-lo hematopoietic stem cell. Cell 44(4): 653–62.
- Ng E et al. (2024) Long-term engrafting multilineage hematopoietic cells differentiated from human induced pluripotent stem cells. Nature Biotechnology Epub ahead of print, DOI: 10.1038/s41587-024-02360-7.
- Naik SH et al. (2014) Cellular barcoding: A technical appraisal. Experimental Hematology 42(8): 598–608
- Notta F et al. (2011) Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 333(6039): 218–21.
- Okada S et al. (1992) In vivo and in vitro stem cell function of c-kit- and Sca-1-positive murine hematopoietic cells. Blood 80(12): 3044–50.
- Osawa M et al. (1996) In vivo self-renewal of c-Kit+ Sca-1+ Lin(low/-) hematopoietic stem cells. J Immunol 156(9): 3207–14.
- Page KM et al. (2011) Total colony-forming units are a strong, independent predictor of neutrophil and platelet engraftment after unrelated umbilical cord blood transplantation: a single-center analysis of 435 cord blood transplants. Biol Blood Marrow Transplant 17(9): 1362–74.
- Peled T et al. (2012) Nicotinamide, a SIRT1 inhibitor, inhibits differentiation and facilitates expansion of hematopoietic progenitor cells with enhanced bone marrow homing and engraftment. Exp Hematol 40(4): 342-55.e1.
- Pellegrin S et al. (2021) Towards manufactured red blood cells for the treatment of inherited anemia. Haematologica 106(9): 2304–11.
- Piau O et al. (2023) Generation of transgene-free hematopoietic stem cells from human induced pluripotent stem cells. Cell Stem Cell 30: 1610–23.
- Prasad VK et al. (2008) Unrelated donor umbilical cord blood transplantation for inherited metabolic disorders in 159 pediatric patients from a single center: influence of cellular composition of the graft on transplantation outcomes. Blood 112(7): 2979–89.
- Purton LE et al. (2000) All-trans retinoic acid enhances the long-term repopulating activity of cultured hematopoietic stem cells. Blood 95(2): 470–7.
- Rai R et al. (2023) An improved medium formulation for efficient ex vivo gene editing, expansion and engraftment of hematopoietic stem and progenitor cells. Mol. Ther. 29: 58–69.
- Sender R & Milo R. (2021) The distribution of cellular turnover in the human body. Nat Med 27: 45–8.
- Shoulars K et al. (2016) Development and validation of a rapid, aldehyde dehydrogenase bright-based, cord blood potency assay. Blood 27(19): 2346–54.
- Shukla S et al. (2017) Progenitor T-cell differentiation from hematopoietic stem cells using Delta-like-4 and VCAM-1. Nat Methods 14(5):, 531–8.
- Shultz LD et al. (2005) Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hematopoietic stem cells. J Immunol 174(10): 6477–89.
- Spangrude GJ & Brooks DM. (1992) Phenotypic analysis of mouse hematopoietic stem cells shows a Thy-1-negative subset. Blood 80(8): 1957–64.
- Spangrude GJ & Brooks DM. (1993) Mouse strain variability in the expression of the hematopoietic stem cell antigen Ly-6A/E by bone marrow cells. Blood 82(11): 3327–32.
- Spangrude GJ et al. (1988) Purification and characterization of mouse hematopoietic stem cells. Science 241(4861): 58–62.
- Storms RW et al. (2005) Distinct hematopoietic progenitor compartments are delineated by the expression of aldehyde dehydrogenase and CD34. Blood 106(1): 95–102.
- Storms RW et al. (1999) Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci USA 96(16): 9118–23.
- Strauss LC et al. (1986) Antigenic analysis of hematopoiesis. V. Characterization of My-10 antigen expression by normal lymphohematopoietic progenitor cells. Exp Hematol 14(9): 878–86.
- Subramaniam A et al. (2020) Lysine-specific demethylase 1A restricts ex vivo propagation of human HSCs and is a target of UM171. Blood 136 (19): 2151–61.
- Sugimura J et al. (2017) Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545(7655): 432–8.
- Suzuki N et al. (2013). Generation of engraftable hematopoietic stem cells from induced pluripotent stem cells by way of teratoma formation. Mol Ther 21(7):1424–31.
- Szilvassy SJ et al. (1990) Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc Natl Acad Sci USA 87(22): 8736–40.
- Szilvassy SJ et al. (2002) Quantitation of murine and human hematopoietic stem cells by limiting-dilution analysis in competitively repopulated hosts. Methods Mol Med 63: 167–87.
- Terstappen LW et al. (1991) Sequential generations of hematopoietic colonies derived from single non lineage-committed CD34+CD38- progenitor cells. Blood 77(6): 1218–27.
- Uchida N et al. (2003) Different in vivo repopulating activities of purified hematopoietic stem cells before and after being stimulated to divide in vitro with the same kinetics. Exp Hematol 31(12): 1338–47.
- Uchida N & Weissman IL. (1992) Searching for hematopoietic stem cells: evidence that Thy-1.1lo Lin- Sca-1+ cells are the only stem cells in C57BL/Ka-Thy-1.1 bone marrow. J Exp Med 175(1): 175–84.
- Velier M et al. (2019) Validation of a semi automatic device to standardize quantification of Colony-Forming Unit (CFU) on hematopoietic stem cell products. Cytotherapy 3: 1–4.
- Vogel W et al. (2000) Clinical applications of CD34(+) peripheral blood progenitor cells (PBPC). Stem Cells 18(2): 87–92.
- Wagner JE et al. (2014) One-unit versus two-unit cord-blood transplantation for hematologic cancers. N Engl J Med 371(18): 1685–94.
- Wang M et al. (2022) Genotoxic aldehydes in the hematopoietic system. Blood 139(14): 2119–29.
- Whitlock CA & Witte ON. (1982) Long-term culture of B lymphocytes and their precursors from murine bone marrow Proc Natl Acad Sci USA 79(11): 3608–12.
- Yang H et al. (2005) Association of post-thaw viable CD34+ cells and CFU-GM with time to hematopoietic engraftment. Bone Marrow Transplant 35(9): 881–7.
- Yoo KH et al. (2007) The impact of post-thaw colony-forming units- granulocyte/macrophage on engraftment following unrelated cord blood transplantation in pediatric recipients. Bone Marrow Transplant 39(9): 515–21.