B Cell Isolation for More Sensitive Array-Based CLL Assays
- Document # 27196
- Version 1.2.0
- Dec 2024
Introduction
B cell chronic lymphocytic leukemia (CLL) is the most common type of leukemia in the world. CLL affects B cell lymphocytes and causes malignant cells to proliferate and accumulate in the bone marrow and peripheral blood.1,2 These malignant CLL cells are distinguished from normal B lymphocytes by the expression of the CD5 surface marker, in addition to CD19, CD20, and CD23.1 They also carry genetic abnormalities, which can differ among subtypes of CLL and among different leukemic subpopulations in the same individual.3,4
CLL is a highly variable disease and can be divided into a number of subtypes based on the distribution of genetic abnormalities present in the malignant cell population. Since these abnormalities can indicate the course that the disease will take, it is essential that CLL assays are able to identify all underlying mutations with the highest-possible sensitivity and accuracy.5,6 In this document, we discuss the advantages of B cell isolation in increasing the sensitivity of array-based CLL assays.
Array-Based Assays for CLL
Array-based comparative genomic hybridization (array-CGH) and array-based single-nucleotide polymorphism analysis (array-SNP) are powerful, efficient, and increasingly common tools for profiling genetic abnormalities in CLL and other hematological malignancies.7-9 Array-based CLL assays have several advantages over older cytogenetic techniques, such as fluorescence in situ hybridization (FISH). For example, it is possible to screen tens of thousands of loci in parallel using arrays, whereas FISH is typically limited to a few probes. Similarly, arrays allow mutations to be detected at single-nucleotide resolution, whereas FISH and conventional cytogenetic techniques can detect only gross chromosomal mutations. Recent research shows that array-based assays can detect abnormalities missed by FISH.9-14 However, the sensitivity of these arrays is contingent upon starting with a purified cell population and, thus, upstream sample preparation.
B Cell Isolation for Increased Cytogenomic Assay Sensitivity
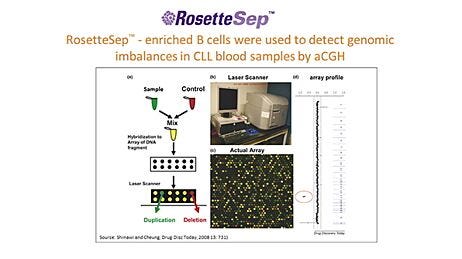
The purpose of molecular assays for CLL is to detect or quantify a specific DNA sequence or RNA transcript in the malignant B cell population. Therefore, the sensitivity and accuracy of the assay increases with the proportion of B cells in the cell population used for DNA/RNA extraction. It has been shown that enriching blood samples for B cells improves the sensitivity of molecular assays because it reduces the effects of normal clone contamination caused by DNA from non-B cells.15 Furthermore, B cell enrichment allows more appropriate comparisons of log2 ratios among samples by eliminating variability caused by differences in B cell frequency.9,16 For example, Hagenkord & Chang (2009) compared array-SNP results on a sample before and after B cell enrichment using RosetteSep™, a reagent for isolating cells directly from whole blood. They showed that enriching B cells before array-SNP allows aberrations that would otherwise have been missed to be detected. (Figure 1).9

Figure 1. 13q14 Regional Copy Number Heterogeneity and Efficacy of B Cell Enrichment
(A) SNP array-based karyotype of the 13q14 locus of a CLL sample with 17% CD5+/CD19+ CLL cells by flow cytometry. (B) Array-based karyotype of the same sample as (A), but enriched for B cells before DNA extraction, revealing a 13q14 aberration that appears partially heterozygously deleted (light blue) and partially homozygously deleted (dark blue). Dark blue indicates copy number of zero, light blue indicates copy number of one, and yellow indicates copy number of two. The log2 ratio plot is shown as a smoothed average over 10 SNPs. Figure and caption adapted from Hagenkord & Chang (2009) with permission from Macmillan Publishers Ltd.
In another publication, Hagenkord et al. (2010) used array-SNP to analyze 6 paired samples that were either enriched for B cells using RosetteSep™ or left unenriched. They found that array-SNP detected lesions that would have been missed by FISH and noted the importance of B cell enrichment as a sample preparation step, particularly for samples with a low tumor burden.16
Virtually all tumor samples contain some ‘normal clone contamination’ from non-neoplastic cells. This will dampen the signals from the tumor and cause failure of both copy number and genotyping algorithms… In our hands, enrichment for B cells not only evens the playing field with regard to comparing log2 ratios between samples, but also resolves lesions in samples with low tumor burden or small subclones.
As a final example, Patel et al. (2008) compared array-CGH using RosetteSep™-enriched CLL samples to FISH using non-enriched samples.17 In several cases, array-CGH detected aberrations that were missed by FISH (Table 1). Furthermore, Patel et al. noted that their assay was twice as sensitive as previous studies had reported, which they attributed to enriching B cells prior to DNA extraction. These studies highlight the benefits of B cell isolation in improving the sensitivity of molecular assays for CLL detection.
| Abnormalies Detected | ||
|---|---|---|
| Case Number | FISH | Array-CGH |
| 25 | 34 | 35 |
| 6 | 0 | 2 |
| 31 | 34 | 37 |
Data from Patel et al. 2008
…our clonal detection cutoff value of 25% is two times more sensitive than what has been previously reported. This increase in sensitivity is most likely due to using DNA from an enriched population of cells in our study. Therefore, these results suggest that for CLL samples, enriching for B-cells allows for higher sensitivity for detecting clonal abnormalities using array-CGH analysis.
Summary
When the starting frequency of B cells in a sample is low, the sensitivity of array-based assays for CLL detection can be impacted. The publications mentioned above show that incorporating B cell isolation into the sample preparation step can significantly improve the sensitivity and consistency of molecular assays for CLL, thereby ensuring accurate testing results. To facilitate CLL research, STEMCELL Technologies offers efficient and easy-to-use immunomagnetic and immunodensity selection platforms for B cell isolation that can be easily incorporated into existing protocols.

Figure 2. Purity of CD19+ CD5+ CLL Cells Following CLL Isolation
CD19+ CD5+ CLL Cells were isolated by incorporating RosetteSep™ into the density gradient centrifugation step (RS+DGC, purple squares) or were enriched using density gradient centrifugation alone (DGC, grey triangles). Adapted from Essakali et al. 2008, under the terms of the Creative Commons Attribution License.
Fast and Easy Immunomagnetic B Cell Isolation from CLL Samples
EasySep™ is a fast, easy, and column-free system for immunomagnetic cell isolation in as little as 8 minutes. Positive selection protocols can be used to isolate desired cells, while negative selection protocols remove unwanted cells to leave you with untouched B cells for the detection of malignant cells in CLL samples. EasySep™ technology can be used with a variety of downstream applications, including FISH, array-based assays, and next generation sequencing (NGS).
Skip Centrifugation and Isolate Cells Directly from Blood
Obtain untouched, highly purified cells directly from blood without the need for sedimentation or red blood cell lysis using EasySep™ Direct.
To increase sample throughput and free up valuable technician time, EasySep™ technology can be automated using RoboSep™ cell isolation instruments. With true walk-away automation, you can isolate B cells from up to 16 samples at once with as little as 5 minutes of hands-on time.
Minimize Sample Handling by Automating Cell Isolation
Increase your lab’s throughput and ensure technician safety when handling samples of unknown origin by using RoboSep™ cell isolation prior to laboratory assays for CLL detection.
Immunodensity B Cell Separation Directly from Whole Blood
RosetteSep™ offers rapid, high-throughput cell isolation directly from human peripheral blood samples in as little as 25 minutes. By crosslinking unwanted cells to red blood cells (RBCs) present in the sample, target cells are purified during standard density gradient centrifugation. To simplify blood layering and ensure sample consistency when isolating cells, RosetteSep™ can be paired with SepMate™, a specialized tube for density gradient centrifugation, and a density gradient medium such as Lymphoprep™.

The short purification time, the independence from expensive and time-consuming procedures such as FACS and MCS, and the flexible adjustment of cell yields makes [RosetteSep™] an attractive purification method for a wide spectrum of downstream applications… in which a CLL purity of >90% is desirable.
Table 3. Products for Immunodensity-Based Cell Isolation from CLL samples*
STEMCELL offers several products for B cell isolation from CLL samples, ensuring that an ideal platform is available for every application. To complement our existing products, we also offer custom cell separation kits for your specific needs. To inquire about a custom cell isolation kit, contact techsupport@stemcell.com.
*Certain products are only available in select territories. Please contact your local sales representative or the Product & Scientific Support team at techsupport@stemcell.com for further information.
Related Resources
- Chiorazzi N et al. (2005) Chronic lymphocytic leukemia. N Engl J Med 352(8): 804–15.
- Keating, MJ. (1999) Chronic lymphocytic leukemia. Semin Oncol 26(5 Suppl 14): 107–14.
- Dierlamm J et al. (1997) Genetic abnormalities in chronic lymphocytic leukemia and their clinical and prognostic implications. Cancer Genet Cytogenet 94(1): 27–35.
- Kipps TJ. (2008) Genomic complexity in chronic lymphocytic leukemia. Blood 112(5): 1550.
- Juliusson G et al. (1990) Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med 323(11): 720–4.
- Shanafelt TD et al. (2004) Prognosis at diagnosis: integrating molecular biologic insights into clinical practice for patients with CLL. Blood 103(4): 1202–10.
- Simons A et al. (2012) Genome-wide arrays in routine diagnostics of hematological malignancies. Hum Mutat 33(6): 941–8.
- Vermeesch J et al. (2012) Genome-wide arrays: quality criteria and platforms to be used in routine diagnostics. Hum Mutat 33(6): 906–15.
- Hagenkord JM & Chang CC. (2009) The rewards and challenges of array-based karyotyping for clinical oncology applications. Leukemia 23(5): 829–33.
- Gunn SR et al. (2008) Whole-genome scanning by array comparative genomic hybridization as a clinical tool for risk assessment in chronic lymphocytic leukemia. J Mol Diagn 10(5): 442–51.
- Gunn SR et al. (2009) Atypical 11q deletions identified by array CGH may be missed by FISH panels for prognostic markers in chronic lymphocytic leukemia. Leukemia 23(5): 1011–7.
- Pfeifer D et al. (2007) Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood 109(3): 1202–10.
- Lehmann S et al. (2008) Molecular allelokaryotyping of early-stage, untreated chronic lymphocytic leukemia. Cancer 112(6): 1296–305.
- Schwaenen C et al. (2004) Automated array-based genomic profiling in chronic lymphocytic leukemia: development of a clinical tool and discovery of recurrent genomic alterations. PNAS 101(4): 1039–44.
- Garnis C et al. (2005) High-resolution array CGH increases heterogeneity tolerance in the analysis of clinical samples. Genomics 85(6): 790–3.
- Hagenkord JM et al. (2010) Array-based karyotyping for prognostic assessment in chronic lymphocytic leukemia: performance comparison of Affymetrix 10K2.0, 250K Nsp, and SNP6.0 arrays. J Mol Diagnos 12(2): 184–96.
- Patel A et al. (2008) Validation of a targeted DNA microarray for the clinical evaluation of recurrent abnormalities in chronic lymphocytic leukemia. Am J Hematol 83(7): 540–6.
- Essakali S et al. (2008) Negative selection of chronic lymphocytic leukaemia cells using a bifunctional rosette-based antibody cocktail. BMC Biotech 8:6.
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration