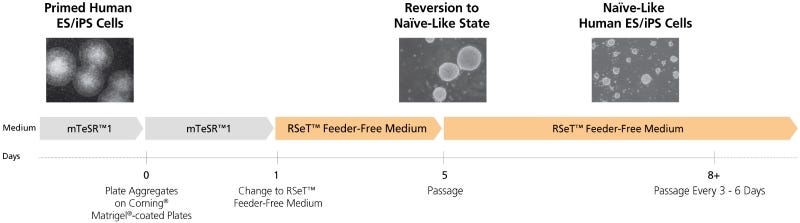
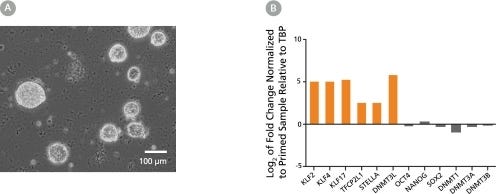
RSeT™ Feeder-Free Medium
Serum-free medium for naïve-like human pluripotent stem cells
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration
Overview
More Information
| Safety Statement | CA WARNING: This product can expose you to Progesterone which is known to the State of California to cause cancer. For more information go to www.P65Warnings.ca.gov |
|---|
Data Figures
Protocols and Documentation
Find supporting information and directions for use in the Product Information Sheet or explore additional protocols below.
Applications
This product is designed for use in the following research area(s) as part of the highlighted workflow stage(s). Explore these workflows to learn more about the other products we offer to support each research area.
Resources and Publications
Educational Materials (3)
Yeda Research and Development Ltd., an Israeli corporation (“YEDA”) is the owner of all right and title to certain patents applications and other proprietary rights which cover (i) the use of the product you are purchasing, such product comprising of media culture compositions to produce novel fully Naïve Induced Pluripotent Stem Cells (“iPSC’s”) (“the Product”) and (ii) the use of the iPSC’s themselves. By purchase of the Product, you shall have, subject to the terms of use below, a limited non-exclusive licence to use the Products to generate iPSC’s and the iPSC’s themselves. YOU HEREBY AGREE TO USE THE PRODUCT AND ANY RESULTS OR PHYSICAL MATTER, INCLUDING IPSC’S, GENERATED FROM THE USE OF THE PRODUCT, OR ANY COPIES OR DERIVATIVES OF ANY OF THE FOREGOING, SOLELY FOR THE PURPOSE OF CONDUCTING NON-COMMERCIAL RESEARCH ACTIVITIES AND PUBLISHING THE RESULTS OF SUCH ACTIVITIES. YOU HEREBY ACKNOWLEDGE THAT ANY USE THEREOF FOR PURPOSES OTHER THAN NON-COMMERCIAL RESEARCH AND PUBLICATION IS PROHIBITED AND SHALL BE PROSECUTED TO THE FULL EXTENT OF THE LAW. IN THE EVENT YOU WISH TO USE THE ABOVE FOR ANY OTHER PURPOSE, A SEPARATE LICENSE IS REQUIRED. YOU CAN CONTACT YEDA AT INFO.YEDA@WEIZMANN.AC.IL FOR THE PURPOSE OF OBTAINING A SEPARATE LICENCE IN THIS REGARD, YEDA BEING IN NO WAY OBLIGATED TO ENTER INTO OR NEGOTIATE SUCH A LICENCE WITH YOU. Nothing contained herein shall be deemed to be a representation or warranty, express or implied, by YEDA that patent applications relating to the Product and the iPSC’s will be granted; (ii) that patents obtained on any of the said patent applications are or will be valid or will afford proper protection; (iii) that the use of the Product or the iPSC’s will not infringe the rights of any third party; or (iv) as to the potential, use, exploitability, merchantability, suitability, effectiveness, fitness for purpose or safety of the Product and the iPSC’s.Under no circumstances shall YEDA be liable for any claims, demands, liabilities, costs, losses, damages or expenses (including legal costs and attorney's fees) of whatever kind (any of the above, a “Loss”) caused to or suffered by any person or entity that directly or indirectly arise out of the use of the Product or the iPSC’s. Copyright © 2017 by STEMCELL Technologies Inc. All rights reserved including graphics and images. STEMCELL Technologies & Design, STEMCELL Shield Design, Scientists Helping Scientists, STEMdiff, and RSeT are trademarks of STEMCELL Technologies Canada Inc. mTeSR is a trademark of WARF. All other trademarks are the property of their respective holders. While STEMCELL has made all reasonable efforts to ensure that the information provided by STEMCELL and its suppliers is correct, it makes no warranties or representations as to the accuracy or completeness of such information.
PRODUCTS ARE FOR RESEARCH USE ONLY AND NOT INTENDED FOR HUMAN OR ANIMAL DIAGNOSTIC OR THERAPEUTIC USES UNLESS OTHERWISE STATED. FOR ADDITIONAL INFORMATION ON QUALITY AT STEMCELL, REFER TO WWW.STEMCELL.COM/COMPLIANCE.
CA WARNING: This product can expose you to Progesterone which is known to the State of California to cause cancer. For more information go to www.P65Warnings.ca.gov