The Predictive Power of Organoids in Drug Discovery
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration
- Document # MR27253
- Version 1.0.0
- Feb 2024
Introduction
There is a pressing need to improve the efficiency of the drug development process and reduce drug attrition rates. Decisions at each stage of the process impact the likelihood of success in bringing new therapeutics into the clinic. Consequently, drug developers need a thorough understanding of how a therapeutic candidate performs in human-relevant assays before advancing these candidates into latter, resource-intensive clinical phases. Organoids can address this need by enabling drug developers to closely model human biology for a comprehensive profile of potential therapeutics’ toxicity, efficacy, and mechanism of action. Biologically relevant, in vitro, stem cell-based organoid models can provide early insights to make informed and confident predictions about the clinical performance of a prospective therapeutic agent.
Read through this overview to understand how the drug discovery landscape is evolving and how organoid-based approaches are being incorporated in drug evaluation and toxicology studies to add predictive value. Best practices and standardization efforts that can maximize the power of organoids to yield reliable, reproducible, and biologically relevant results are also discussed.
A Need for Biological Relevance: Snapshot of the Drug Discovery Landscape
Bringing new, effective therapeutics to market is a long, expensive, and high-risk process that can take over 15 years with an average cost of $1 - 2 billion for each new drug to be approved for clinical use. Moreover, the overall success rate of transitioning from drug discovery through to market approval is less than 10%.1 Failure in phase II and phase III clinical trials is a major driver of cost and inefficiencies in the drug development process; approximately 50% of these failures are caused by inadequate drug efficacy and 15 - 25% by safety concerns.2,3
Many approaches are used to identify and validate biological targets and then to identify and optimize therapeutic candidates; biological assays using 2D immortalized cell cultures and non-human animal species are common methods. While 2D immortalized cell cultures allow for high-throughput screening at low cost and animal studies enable investigation of potential therapeutic effects in an in vivo setting, both classes of conventional model systems have limitations in predicting human responses to drugs. There is a recognized need for models with a higher predictive value that can be used to evaluate the performance of candidates in a human-relevant setting.
New approach methods (NAMs, alternatively termed “non-animal models” or “new assay modalities”) represent innovative approaches for scientific research that aim to reduce or replace the use of animals in experiments and allow for rapid and effective risk assessment. These methodologies may include the use of cell-based assays, tissue engineering techniques, computational models, and exposure prediction approaches. Cell-based assays can include high-throughput screening and omics technologies to understand molecular processes underlying the complex disease etiology or the mode of action of a therapeutic candidate.
As therapeutic strategies continue to diversify and more complex diseases are studied, there is growing recognition that immortalized cell lines and animal models are not sufficient to gather accurate insights into disease biology and treatment responses. There is also pressure to improve the productivity of the drug development process and shorten the time to market for a new drug for patients in need. Moreover, researchers are proactively seeking and implementing NAMs to improve their workflows for sustained and long-term success in their respective drug discovery programs. Drug developers are also adopting NAMs to address the financial pressures associated with the costly drug development process. With NAMs, there is renewed potential to ensure return on investment while supporting the desire of researchers and drug developers to bring safe and effective therapies to market as quickly as possible.
Several regulatory jurisdictions have passed legislation supporting the development and timely incorporation of scientifically validated NAMs to replace, reduce, or refine the use of vertebrate animals in the testing and evaluation of substances. For example, in 2022 the U.S. passed the FDA Modernization Act 2.0, which provides more clarity on the acceptability of in vitro tests, making it explicit that they are accepted in lieu of animal studies as long as data submitted proves they are equally capable of assessing risk. Other examples of such shifts include Bill S-5 passed by the Government of Canada and the EU’s commitment to modernize science by reducing animal testing and supporting alternative testing methods. Indeed, the existing legislative framework in the EU already sets strict rules and conditions on how animal testing can be undertaken where still necessary with the goal of fully phasing out all animal use in research and for regulatory purposes in the EU.
Among NAMs, organoids provide an opportunity to improve in vitro-to-human translation and facilitate progress toward the replacement of animals used in biomedical research. A number of drug discovery projects are now utilizing organoids, stem cell-based in vitro models, for improved internal decision-making at multiple steps in the drug development process and are showing progress in increasing the chances of clinical success.4-6 Furthermore, cross-firm and cross-sector collaborative groups have been established to expedite advances in testing methods by promoting the sharing of information and resources around emerging technologies, including organoids, for easier adoption and increased standardization. These groups include the IQ Consortium, IATA Case Studies Consortia, Investigative Toxicology Leaders Forum,7 National Center for Advancing Translational Sciences, and Organoid Task Force of the American Society for Cell Biology.
Below, we will review how organoid-based approaches add biological relevance while ensuring scalability and reproducibility to address specific needs along the process of drug development.

Figure 1. Advancing Therapeutic Development with Human-Relevant Model Systems
The therapeutic development process holds great potential for improvements in productivity. The typical program involves several biological experimental systems to (1) identify and validate druggable targets, (2) identify and optimize lead candidates, and (3) further assess preclinical candidates for their human-relevant responses as accurately as possible before proceeding with the clinical phase of drug development. Advances in tissue modeling techniques are paving the path for improvements in how scientists develop treatments for disease management. Although 2D immortalized cell lines and animal models are used in multiple domains of biomedical research, they lack biological relevance to the human setting. However, with continued progress in therapeutic strategies to tackle multifaceted, complex diseases, there is a growing need to incorporate more human-relevant disease and healthy tissue models to uncover risks associated with advancing a potential treatment to the clinic. Organoid models of human disease and healthy human tissue can help evaluate efficacy and predict toxicity, respectively.
Human Stem Cell-Based In Vitro Systems: Overview of Organoids
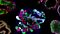
Organoids are 3D cell cultures that incorporate some of the key features of the represented organ. These in vitro culture systems are characterized by the self-organization of multiple, organ-specific cell types into a spatial arrangement similar to what is observed in vivo and are capable of recapitulating some functions of the represented organ.8,9
Organoids may be generated from tissue sample-derived adult stem cells or via the directed differentiation of pluripotent stem cells (PSCs), which can be embryonic (ESCs) or induced from other human cell types (iPSCs) (Figure 2).8,9 Some organoids derived from tissue stem cells (TSCs) or PSCs can be expanded and maintained over several passages, generating up to thousands of organoids in one well of a multi-well culture plate, depending on the tissue type. Thus, organoids provide a highly robust and scalable platform for researchers to perform human-relevant assays and study a wide range of subjects.
To initiate organoids, PSCs or TSCs are cultured in media containing specific growth factors that mimic their in vivo niche. Some organoids require the presence of additional extracellular matrix for optimal growth. Exposure to these conditions directs the embedded cells to follow their innate developmental programs and self-organize into 3D organoid structures that closely model the intracellular and intercellular mechanisms observed in vivo. Organoid cultures have shown remarkable genetic stability during passaging; whole genome sequencing of liver organoids clonally expanded from a single liver progenitor cell revealed only one synonymous base substitution after three months in culture.10 To date, organoid cultures have been described for a variety of tissues, including intestine, liver, pancreas, kidney, lung, and brain tissues.

Figure 2. Tissue-Modeling Organoids Can Be Derived from Human TSCs or PSCs
TSC- and PSC-derived organoids are generated from stem cell populations that undergo differentiation pathways to produce 3D in vitro cultures containing multiple tissue-specific cell types interacting with each other, representative of functional tissue. TSC-derived organoids are often derived from biopsy samples or surgically resected tissue. PSC-derived organoids can be generated from ESCs or iPSCs that have the ability to differentiate into any cell type of the body. The unique advantage of organoids is their ability to capture genetic and phenotypic characteristics of the original tissue in 3D in vitro cultures, providing a more representative profile of cellular diversity, gene expression, and cellular function.
Organoids in Drug Discovery
Since the early work on organoids over a decade ago (as discussed in this e-book on organoid research), the development of organoid culture techniques has provided researchers with new experimental platforms for studying normal and pathological biology. Many drug development pipelines, however, still rely on 2D cell culture and animal models to provide necessary non-clinical insights. Various reasons motivate these choices, including ease of use, the existence of established assays, and the availability of historical benchmarks. While these conventional approaches continue to be invaluable for therapeutic discovery and development, they are limited in providing the predictivity required to reduce late-stage attrition, especially in the cases of solid cancers and complex diseases. First, the interspecies genetic and physiological differences between humans and animals limit the power of animal studies to predict human-relevant toxicity mechanisms and disease processes. Second, immortalized cell lines can only model metabolic or transport activity for their specific cell type rather than exemplifying a more comprehensive set of biological characteristics of the functional organ. With the development of human organoid culture systems, there is new potential to bring human-specific mechanistic insights into decision-making processes prior to clinical studies.
Features that make organoid culture systems valuable and predictive models for therapeutic discovery
- Species-Specific and Patient-Specific Models: Organoids derived from primary human cells make them highly representative of human physiology and avoid interspecies variability. Normal or disease-representative organoids can be generated from any individual. PSC-derived organoids can also be generated from patient skin, blood, hair, or urine samples, which is especially useful for organs that are difficult to biopsy. This allows researchers to study disease mechanisms and test potential therapeutic candidates in a population segment- and disease-specific context.
- High-Fidelity Biological Models: Organoids derived from unaltered human cells (i.e. wild-type cells) are genetically identical to the patient/tissue of the original sample. Capable of recapitulating tissue-specific cell composition, enzyme and transporter profiles, and physical characteristics, this model system helps predict the impacts of treatment on human tissues.
- Expandable and Bankable Cultures: Organoids can be derived from minimal amounts of normal or diseased tissue biopsies and can be expanded for several passages, enabling the generation of living biobanks, an important resource in biomedical research; for instance, intestinal organoid biobanks have facilitated research in colon cancer11 and cystic fibrosis.12 Inter-donor variability can be managed and understood through donor standardization and representative panels as needed. Patient-derived organoids (PDOs) have been generated by designated biobanks, such as the Hubrecht Organoid Technology (HUB) Biobank, UZ/KU Leuven Biobank, and Discover Together Biobank. Additionally, iPSC technology enables the banking of patient-derived stem cells. Accordingly, there are a growing number of well-characterized iPSC biobanks, such as the Coriell Institute for Medical Research and WiCell Research Institute, and commercially available iPSC lines.
- Versatility in Assay Design: Organoid-derived cells are amenable to genetic modifications and are compatible with assays in a wide variety of formats, including dome-embedded, suspension, air-liquid interface, high-throughput 96/384-well formats, and organ-on-a-chip (OoC)/microphysiological systems (MPS).
Optimizing Organoid Models to Better Predict Clinical Outcomes
In this webinar, Dr. Sylvia Boj (HUB Organoids) and Dr. Martin Stahl (STEMCELL Technologies) discuss the products, protocols, and assays that enable the use of organoid models in drug discovery research, with a focus on intestinal and liver organoids.
Watch Now >Performance of Organoids in Drug Evaluation Studies
Organoids are being used in a range of cell-based assays to assess the human drug response. Several studies have demonstrated the performance of organoid-based platforms in testing drug efficacy and toxicity:
- Patient-derived tumor organoids predicted treatment response in more than 80% of metastatic colorectal cancer patients treated with irinotecan-based chemotherapy (accuracy: 80%).13
- A hepatic organoid-based high-throughput toxicity screen in 384-well plates tested 238 marketed drugs through multiplexed readouts measuring cell viability, bile acid transport activity, and mitochondrial toxicity with high predictive values (sensitivity: 88.7%, specificity: 88.9%).14
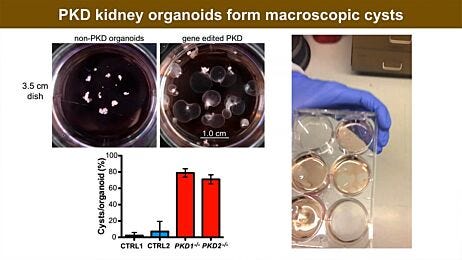
- A kidney organoid-based high-throughput platform in 384-well plates accurately predicted cisplatin-induced nephrotoxic effects in a dose-dependent manner through a luminescence-based cell viability assay and biomarker expression analysis.15
- Patient-derived rectal cancer organoids predicted response to chemoradiation through organoid size as the readout (accuracy: 84.43%, sensitivity: 78.01%, specificity: 91.97%).16
- Patient-derived organoids derived from metastatic gastrointestinal cancers predicted patient response to targeted agents or chemotherapy in clinical trials (sensitivity: 100%, specificity: 93%).17
Thus, organoids have demonstrated promise in drug evaluation studies, showcasing notable specificity and sensitivity. This predictive power, coupled with their ability to model patient-specific responses, positions organoids as a powerful tool for advancing personalized medicine and streamlining the therapeutic development process.
Organoids in Adverse Outcome Pathways
Evaluating adverse outcome pathways (AOPs) is a useful approach to predict clinical outcomes by gathering information on key biological processes that lead to a certain undesirable outcome (Figure 3). Organoids have the potential to measure key events in established AOPs and therefore provide predictive and mechanistic insights into potential adverse effects of therapies in development. The following studies have utilized organoids to gain insights into a multi-event path of causality by assaying individual events to predict a health outcome:
- Lung, liver, and kidney organoid-based models were used in several SARS-CoV-2-related studies to understand viral pathogenesis and potential antiviral treatments. These studies mapped organoid-based data, including gene expression and cell behavior, to AOP frameworks to predict SARS-CoV-2-related in vivo effects and then validated these findings by comparison with COVID-19 patient data.18-26
- Liver organoids derived from human pluripotent stem cells (hPSCs) were used as a model to study microplastic toxicity. The mechanisms of polystyrene microplastic-induced hepatotoxicity and lipotoxicity were determined by measuring several causally linked key events, including lipid metabolism, oxidative stress, and inflammation response. The combined application of liver organoids with specific in vitro assays allowed for the collection of predictive data along the AOPs (i.e. AOP 36), providing new insights to inform the risk assessment of microplastics.27
By anchoring organoid-based data to the AOP mechanistic framework, researchers can advance evidence-based, mechanistic toxicology and improve the predictivity of clinical outcomes while reducing their reliance on animal testing.

Figure 3. Organoids Can Provide Mechanistic Insights Along the Adverse Outcome Pathway Framework
An AOP starts with the molecular initiating event (MIE), which describes how a chemical interacts with a biological target like DNA, which will then lead to a sequential chain of key events (KEs). Each KE is an alteration of biological processes. The AOP framework provides mechanistic information required to increase confidence in decision-making and can help promote the use of non-animal methods to predict the toxicity of a lead candidate. By using AOPs as a conceptual framework, results generated by different approaches can be related to KEs along the AOP pathway and then linked to an adverse outcome. In vitro, organoid-based assays can provide biologically relevant evidence for one or more KEs to help mediate the prediction of adverse outcomes.
Maximizing Biological Relevance: Organoids and Complex In Vitro Systems
Organoid co-cultures and organ-on-a-chip (OoC) systems are advanced in vitro models that further enhance the physiological relevance of organoids by incorporating a broader array of tissue components compared to conventional organoid cultures. In particular, TSC-derived organoids are composed mainly of cell types found in the epithelium. In contrast, organoid co-culture systems allow researchers to study more intercellular interactions than possible with organoid cultures alone by culturing diverse tissue cell types of an organ, such as epithelial and immune cell types, together in a controlled environment to better simulate in vivo organization. OoCs, on the other hand, use microfluidic technology to recreate the physiological conditions of specific organs on a miniature scale, enabling researchers to add mechanical forces and fluid flow. Leveraging organoids as a biological component of OoCs is recognized as a powerful approach to optimize human biological relevance.28,29 By incorporating organoids into these systems, researchers can better understand the dynamic functions and responses of organs for drug evaluation and disease modeling studies.
The following are some examples where organoid co-cultures and OoC technologies have helped answer specific biological questions:
- Gut Immunology: Intestinal epithelial organoids can be co-cultured with enteric pathogens and/or immune cells to address research questions around intestinal barrier function and mucosal immunology.30-35
- Respiratory Tract Infections: The respiratory tract is a major site of entry for many pathogenic organisms. Organoids modeling the lungs, airways, and alveoli have been developed and can be co-cultured with immune cells to investigate host immune responses.36-38
- Cancer Cell-Immune Interactions: Cancer cell-immune interactions play a major role in shaping tumor progression and treatment response. Patient-derived cancer cell organoids can be co-cultured with immune cells to address research questions around immune evasion mechanisms and devise effective immunotherapeutic strategies.39
- Neuroinflammation: In many pathological conditions of the central nervous system, there is often an inflammatory response in the affected areas of the brain or spinal cord. Organoid co-cultures combining neurons, astrocytes, and microglia can enable the study of complex cross-talk between these cell types, offering insights into neuroinflammatory responses and the pathophysiology of various diseases.40
- Tissue Modeling: OoCs can recapitulate the intricate microenvironment of a functional healthy or diseased tissue. 41,42 Assay-ready commercial OoC systems can incorporate organoid-derived cells for optimized biological characteristics that mimic the original properties of the donor, including specific disease processes, and can be maintained in cultures in vitro.
- Podcast Episode “Organ-on-a-Chip” Featuring Dr. Bas Trietsch
- Multi-Organ Interactions/Systemic Effects: Organoid lines representing different organs interconnected on a microfluidic chip offer a platform to mimic the potential interaction of an organ with at least one other organ. For instance, integrated organoid-based organ-on-a-chip systems of the liver, heart, lungs, vascular endothelium, brain, and testes have been used to study drug responses against capecitabine and ifosfamide.43
Overall, these complex in vitro systems provide more biologically relevant and sophisticated platforms, enabling researchers to supplement current drug evaluation studies for a better understanding of dynamic intercellular and inter-tissue interactions that influence human-relevant drug responses.
Beyond Limitations: Solutions to Optimize Organoid-Based Models
Generally, organoids lack components like blood vessels and immune cells and do not fully replicate the complexity of tissues found in vivo. As discussed in the previous section, more complex intercellular and inter-tissue interactions can be modeled through organoid co-cultures and OoC systems. Advancements in organoid co-culture systems have enabled the incorporation of multiple cell types, such as immune cells or endothelial cells, into organoid cultures.30-40 In neuroscience research, interactions between multiple brain regions or multiple tissues are studied by developing multi-lineage assembloids, in which cells migrate or extend projections from one organoid to the next, establish connections, and build neural circuits that model complex cellular interactions of otherwise inaccessible organs. For instance, assembloids have been used to model interactions between different brain regions,44 and co-cultures of human glial cells and cortical organoids have been used to model and study the mechanisms of Alzheimer’s disease and other neurodegenerative diseases.45,46 Researchers have also been successfully incorporating organoids in OoC systems to study the influence of biophysical factors, such as fluid flow, mechanical forces, and nutrient gradients, on tissue function and drug activity.41,42
Although organoid heterogeneity is more biologically relevant than more homogeneous culture systems in many cases, it can present challenges for experimental reproducibility and data analysis. Heterogeneity (for example, different sizes or morphologies) exists between organoids in a single well or dish as well as between organoids and organoid lines from different donors (i.e. donor-to-donor variability). Variability in organoid cultures can also arise from inconsistencies in culture conditions. Standardization of protocols and reagents is key to mitigating this issue, involving precise control over factors like cell isolation procedures and media quality. Quality-controlled, optimized organoid growth and differentiation media can reduce the chances of error in reagent preparation and ensure consistent performance, especially with hard-to-culture lines. Deriving and banking donor-specific iPSC lines and/or organoid lines, and using commercially available iPSC lines can help with standardization in organoid cultures and facilitate efficient collaboration between different institutions and investigators. Thus, advancements in organoid technology with standardized, quality-controlled reagents and designated biobanks are mitigating the limitations due to variability and enabling reliable and scalable workflows.
Ethical and legal considerations are also associated with human-derived organoids, particularly when sourced from stem cells or tissues with unclear consent procedures. Obtaining informed consent and maintaining transparency with donors is one approach to help address ethical concerns and ensure responsible research practices. When possible, leveraging registered commercially available iPSC lines, iPSC biobanks, and/or organoid biobanks is another option to ensure that ethical and legal standards are met.
Best Practices and Standardization Efforts
The Standards for Human Stem Cell Use in Research by the International Society for Stem Cell Research is a rich resource of recommendations that establish the minimum characterization and reporting criteria for working with human stem cells. These guidelines aim to help researchers produce more reproducible, higher-quality results and follow more standardized practices. Another online resource, hPSCreg, provides standardized information about human PSC lines, including the details of their origin, genetic characteristics, and differentiation potential. By collaborating with cell banks and international initiatives worldwide, hPSCreg facilitates collaboration and ensures compliance with ethical and legal considerations. As noted earlier, several cross-firm collaborative groups are also laying down the foundations for best practices and striving toward increased standardization by enabling collaboration and sharing of knowledge and resources around human-relevant in vitro systems. Furthermore, opting for commercially available reagents and published protocols is a convenient way of ensuring that the most optimized and standardized approaches are followed for quality in vitro experiments.
To conclude, organoid-based models hold immense potential in therapeutic discovery and development. By addressing their associated limitations through technological advancements and collaboration, we can fully unlock their capabilities for studying patient-specific disease processes and drug responses.
Key Takeaways
- Although immortalized cell lines and animal models are useful for many applications in drug discovery, they are limited by their reductive and cross-species characteristics, respectively, which affect their ability to predict human in vivo responses with sufficient accuracy.
- Organoids are high-fidelity biological models that can recapitulate the cellular structure and function of specific organs, can be expanded and stored for long periods, and are compatible with a wide range of assays, including high-throughput screening.
- Organoids have demonstrated predictive power with notable sensitivity and specificity in recent drug evaluation studies.
- Several collaborative groups are addressing the need for standardization of new methods in specific contexts of use to effectively apply these methods to improve the efficiency of drug development programs. Partly because of these initiatives, biobanks and commercialized iPSC lines are available to ensure reliable and quality organoid-based research.
- Incorporating human stem cells or stem cell-derived organoids into complex systems such as co-culture and OoC systems is a powerful way to closely model metabolic, transport, and other physiological cellular characteristics of the tissue of interest, maximizing the predictive value of these systems for drug discovery research.
Related Resources
References
- Hinkson IV et al. (2020) Accelerating therapeutics for opportunities in medicine: a paradigm shift in drug discovery. Front Pharmacol 11: 770.
- Paul SM et al. (2010) How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov 9(3): 203–14.
- Harrison R. (2016) Phase II and phase III failures: 2013–2015. Nat Rev Drug Discov 15: 817–8.
- Morgan P et al. (2018) Impact of a five-dimensional framework on R&D productivity at AstraZeneca. Nat Rev Drug Discov 17: 167–81.
- https://www.roche.com/stories/organoids-in-research-technologies
- Herpers B et al. (2022) Functional patient-derived organoid screenings identify MCLA-158 as a therapeutic EGFR × LGR5 bispecific antibody with efficacy in epithelial tumors. Nat Cancer 3: 418–36.
- Beilmann M et al. (2019) Optimizing drug discovery by investigative toxicology: Current and future trends. ALTEX 36(2): 289–313.
- Lancaster MA & Knoblich JA. (2014) Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 345(6194): 1247125.
- Huch M & Koo B-K. (2015) Modeling mouse and human development using organoid cultures. Development 142(18): 3113–25.
- Huch M et al. (2015) Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160(1–2): 299–312.
- van de Wetering M et al. (2015) Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161: 933–45.
- Geurts MH et al. (2020) CRISPR-based adenine editors correct nonsense mutations in a cystic fibrosis organoid biobank. Cell Stem Cell 26: 503–10.e7.
- Ooft SN et al. (2019) Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med 11: eaay2574.
- Shinozawa T et al. (2021) High-fidelity drug-induced liver injury screen using human pluripotent stem cell-derived organoids. Gastroenterology 160(3): 831–46.e10.
- Czerniecki SM et al. (2018) High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 22(6):929–40.
- Yao Y et al. (2020) Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell 26(1): 17–26.
- Vlachogiannis G et al. (2018) Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359(6378): 920–6.
- Katsura H et al. (2020) Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 27(6): 890–904.
- Han Y et al. (2021) Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589: 270–75.
- Huang J et al. (2020) SARS-CoV-2 infection of pluripotent stem cell-derived human lung alveolar type 2 cells elicits a rapid epithelial intrinsic inflammatory response. Cell Stem Cell 27(6): 962–73.e7.
- Simoneau CR & Ott M. (2020) Modeling multi-organ infection by SARS-CoV-2 using stem cell technology. Cell Stem Cell 27: 859–68.
- Shi R et al. (2020) A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584(7819): 120–4.
- Salahudeen AA et al. (2020) Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588(7839): 670–5.
- Monteil V et al. (2020) Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical grade soluble human ACE2. Cell 181(4): 905–13.e7.
- Yang L et al. (2020) A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell 27(1): 125–36.e7.
- Wysocki J et al. (2021) A novel soluble ACE2 variant with prolonged duration of action neutralizes SARS-CoV-2 infection in human kidney organoids. J Am Soc Nephrol 32(4): 795–803.
- Cheng W et al. (2022) Polystyrene microplastics induce hepatotoxicity and disrupt lipid metabolism in the liver organoids. Sci Total Environ. 806(Pt 1): 150328.
- Fowler S et al. (2020) Microphysiological systems for ADME-related applications: current status and recommendations for system development and characterization. Lab Chip. 20(3): 446–67.
- Peters MF et al. (2020) Developing in vitro assays to transform gastrointestinal safety assessment: potential for microphysiological systems. Lab Chip. 20(7): 1177–90.
- Hammoudi N et al. (2022) Autologous organoid co-culture model reveals T cell-driven epithelial cell death in Crohn's Disease. Front Immunol 13: 1008456.
- Jung KB et al. (2018) Interleukin-2 induces the in vitro maturation of human pluripotent stem cell-derived intestinal organoids. Nat Commun 9(1): 3039.
- Kakni P et al. (2022) A microwell-based intestinal organoid-macrophage co-culture system to study intestinal inflammation. Int J Mol Sci 23(23): 15364.
- Noel G et al. (2017) A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci Rep 7: 45270.
- Lemme-Dumit JM et al. (2022) Epithelial and neutrophil interactions and coordinated response to Shigella in a human intestinal enteroid-neutrophil coculture model. mBio 13(3): e0094422.
- Zhou J et al. (2017) Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci Adv 3(11): eaao4966.
- Zhu N et al. (2020) A novel coronavirus investigating and research team. A novel coronavirus from patients with Pneumonia in China, 2019. N Engl J Med 382(8): 727–33.
- Ronaghan NJ et al. (2022) M1-like, but not M0- or M2-like, macrophages, reduce RSV infection of primary bronchial epithelial cells in a media-dependent fashion. PLoS One 17(10): e0276013.
- Lechner AJ et al. (2017) Recruited monocytes and type 2 immunity promote lung regeneration following pneumonectomy. Cell Stem Cell 21(1): 120–34.e7.
- Dijkstra KK et al. (2018) Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 174(6): 1586–98.e12.
- Goshi N et al. (2020) A primary neural cell culture model to study neuron, astrocyte, and microglia interactions in neuroinflammation. J Neuroinflammation 17(1): 155.
- Baudy AR et al. (2020). Liver microphysiological systems development guidelines for safety risk assessment in the pharmaceutical industry. Lab Chip 20(2): 215–25.
- Peters MF et al. (2020). Developing: In vitro assays to transform gastrointestinal safety assessment: Potential for microphysiological systems. Lab Chip 20(7): 1177–90.
- Rajan SAP et al. (2020) Probing prodrug metabolism and reciprocal toxicity with an integrated and humanized multi-tissue organ-on-a-chip platform. Acta Biomater 106: 124–35.
- Paşca SP. (2019) Assembling human brain organoids. Science 363(6423): 126–7.
- Lin YT et al. (2018) APOE4 causes widespread molecular and cellular alterations associated with Alzheimer's Disease phenotypes in human iPSC-derived brain cell types. Neuron 98(6): 1294.
- Ao Z et al. (2021) Tubular human brain organoids to model microglia-mediated neuroinflammation. Lab Chip 21(14): 2751–62.