How to Identify ALDH-Expressing Cells in Human Breast Tissue
Introduction
The non-pregnant mammary epithelium contains three lineages of epithelial cells: estrogen receptor (ER)-expressing, milk lineage, and basal/myoepithelial. The milk lineage cells are a population of ER- cells present in the gland that will proliferate during pregnancy to generate milk-secreting alveolar cells.10–12 Experiments in mice demonstrate that each of these lineages in the postnatal gland is maintained by their own subpopulation of stem cells.13–17 The ALDEFLUOR™ assay identifies a subset of both mouse and human milk lineage cells (Figure 2) that are highly clonogenic in vitro12,18, have stem-like properties19, and are perceived to be the precursors for basal-like breast cancer.20,21 In some breast tumors, the ALDEFLUOR bright (ALDHbr) subpopulation is enriched for cells that can generate tumors when transplanted into immune-deficient recipient mice.19 The presence of ALDHbr cells in an ER+ human breast tumor is predictive of anti-estrogen treatment failure; this is presumably because ALDHbr cells do not express the estrogen receptor and would be selected for during long-term exposure to anti-estrogen agents.22
Aldehyde dehydrogenases (ALDHs) are a family of cytosolic enzymes that are involved in oxidizing a wide variety of aldehydes to their corresponding acids. The presence of these ALDH enzymes enables primitive cells in a variety of tissues to protect themselves from toxic compounds in their environment1, and in cancer, these enzymes provide resistance to chemotherapeutic agents (e.g. cyclophosphamide).2,3 There are 19 ALDH genes in humans4; this variety in isoforms ensures that there are many different enzymes available to catalyze a wide variety of different substrates. The ALDEFLUOR™ assay system is a method that permits the purification of cells expressing high levels of ALDH enzymes. In this assay, ALDEFLUOR™ is supplied in the form of Bodipy™-aminoacetaldehyde diethyl acetal (BAAA-DA) (Figure 1), which by itself, is not a substrate of ALDH. Therefore, BAAA-DA is dissolved in dimethylsulfoxide (DMSO) and exposed to acid to convert it into Bodipy-aminoacetaldehyde (BAAA), which is a fluorescent substrate for ALDH that freely diffuses into live cells. Cells that express the appropriate ALDH enzymes convert BAAA to Bodipy aminoacetate (BAA), which has a net negative charge and can no longer freely diffuse across the cell membrane. Thus, the amount of BAA fluorescence in cells is proportional to ALDH activity. Upon excitation with a 488 nm blue light laser, cells that have accumulated high levels of BAA will have the bright fluorescence in the green light spectrum. A separate incubation with an excess of the ALDH inhibitor, diethylaminobenzaldehyde (DEAB), is used for background fluorescence control (Figure 2). In the first application of the ALDEFLUOR™ assay, the substrate BAAA was initially perceived to be specific for ALDH1A1, since this is the predominant isoform present in human hematopoietic stem cells.5 However, subsequent studies have demonstrated that the ALDEFLUOR™ substrate BAAA interacts with multiple ALDH isoforms, including ALDH1A2, 1A3, 2, 5A1, 6A1, and 7A1, albeit with varying specificities.6–8 ALDEFLUOR™ is not a substrate for ALDH3A1.9
This protocol describes how to stain dissociated normal human breast tissue with the ALDEFLUOR™ Kit in conjunction with antibodies that are commonly used in mammary epithelial cell research. This protocol can be applied to multiple solid tissues, with the main ALDEFLUOR™ variables being the BAAA concentration, incubation time, and antibody selection. For more details regarding ALDEFLUOR™ assay optimization, please see the Technical Bulletin titled “ALDEFLUOR™ Assay Optimization” (Document #29902).
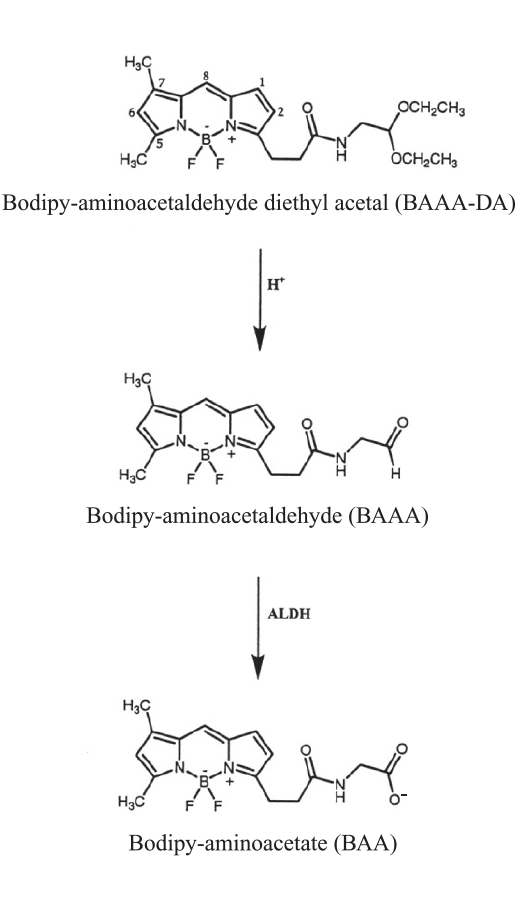
Figure 1. Structure of BAAA-DA, BAAA, and BAA.
Prior to use, Bodipy™-aminoacetaldehyde diethyl acetal (BAAA-DA) is dissolved in DMSO and converted to Bodipy-aminoacetaldehyde (BAAA) when exposed to acid (1 N HCl) for 15 - 30 minutes at room temperature. BAAA can freely diffuse into viable cells and is converted, by ALDH, into Bodipy aminoacetate (BAA), which is negatively charged and retained inside the cells.
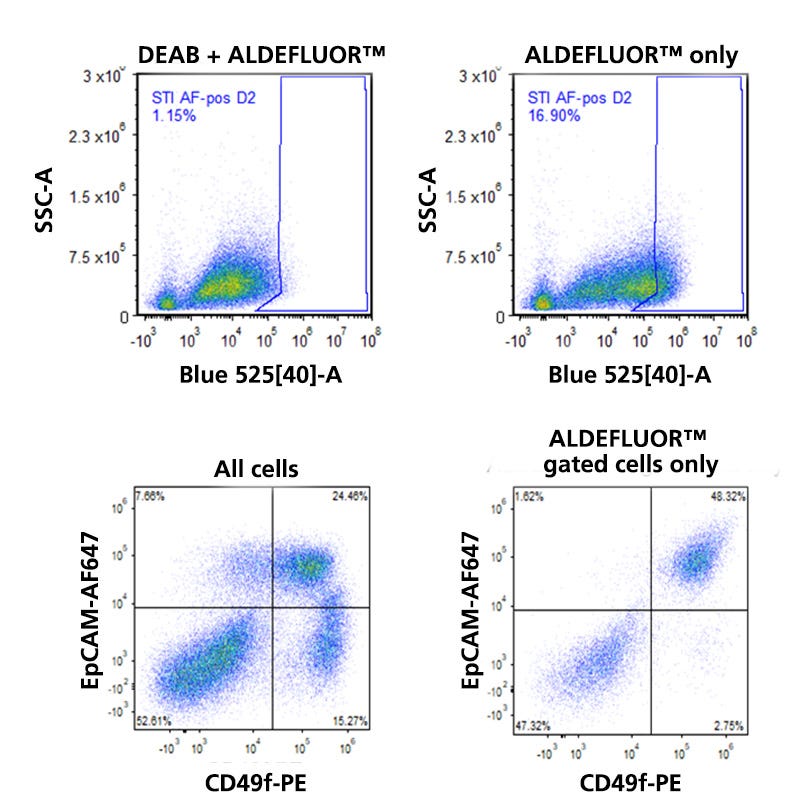
Figure 2. Detection of ALDH Expression in Human Breast Tissue.
Normal human breast tissue from a reduction mammoplasty specimen was dissociated and incubated in the presence of Bodipy™ aminoacetaldehyde (BAAA) with or without the ALDH inhibitor diethylamino-benzaldehyde (DEAB). As illustrated in the flow cytometry dot plots in the top row, the presence of DEAB greatly reduces the frequency of ALDHbr events. The bottom left dot plot shows the distribution of EpCAM and CD49f-expressing cells among all viable CD45- cells. The three main mammary lineages, which are estrogen receptor-expressing (EpCAM+CD49f-), milk lineage (EpCAM+CD49f+), and basal (EpCAMlowCD49f+), are resolvable. Most stromal cells have an EpCAM-CD49f- phenotype. When the mammary cells are gated solely on ALDHbr events, it becomes apparent that high levels of ALDH activity are largely restricted to only a subset of milk lineage and stromal cells.
ALDH Expression in Normal Human Breast Tissue
The following protocol is an example of how to stain normal human breast tissue with the ALDEFLUOR™ Kit and with monoclonal antibodies to detect EpCAM, CD49f, CD45, and CD31 expressing cells for fluorescence-activated cell sorting. The combination of EpCAM and CD49f staining is commonly used to resolve stromal (EpCAM-CD49f-), basal (EpCAM-CD49f+), milk lineage (EpCAM+CD49f+), and estrogen receptor-expressing (EpCAM+CD49f-) cells in the normal mammary epithelium (Figure 2). Antibodies specific to CD45 and CD31 are included to gate out contaminating hematopoietic and endothelial cells, respectively. A viability dye, 4′,6-diamidino-2′-phenylindole dihydrochloride (DAPI), is included to gate out dead cells as these cells can be abundant when dealing with previously cryopreserved samples. The fluorochrome panel chosen for this example is for illustrative purposes only, and the exact fluorochromes used may vary with different flow cytometry instruments; however, in principle, all fluorescent labels that are commonly used in conjunction with other green fluorochromes can be used in combination with ALDEFLUOR™. Please note that the green fluorescence of ALDEFLUOR™-treated cells is very strong, and thus it is important that electronic fluorescence compensations between the green fluorescence channel used to measure ALDH activity and the other fluorescence channels are set accurately using appropriate single color-stained control cells. In the protocol below, we include the presence of BAAA and DEAB in the single color controls that are used to adjust the compensation. At first glance, the inclusion of BAAA and DEAB in these controls is counter-intuitive. However, we recommend this because BAAA, even in the presence of DEAB, dramatically increases the autofluorescence of cells. By including BAAA and DEAB in the single color control tubes, we can more accurately adjust the compensation. The example protocol below assumes that the cells are for analysis only, and as a result, the cells are equally distributed among tubes 1-13. If the cells are for cell sorting, then the bulk (~70%) of them should be allocated to tube 13.
Materials
- ALDEFLUOR™ Kit (Catalog # 01700)
- DMEM/F-12 with 15 mM HEPES (Catalog #36254)
- Tissue Dissociation Flask (Catalog #27300)
- Collagenase/Hyaluronidase (Catalog #07912)
- EpiCult™-C Human Medium Kit (Catalog # 05630)
- Falcon® Conical Tubes, 50 mL (Catalog #38010)
- CryoStor® CS10, 100 mL (Catalog # 07930)
- Trypsin-EDTA (0.25%) (Catalog # 07901)
Protocol
Dissociation of Human Breast Tissue
- Transport human mammary tissue from the operating room on ice in sterile specimen cups, in DMEM/F-12 with 15 mM HEPES supplemented with 5% fetal bovine serum (FBS).
- Transfer tissue to sterile glass Petri dishes, mince with scalpels, and then transfer to tissue dissociation flasks.
- Dilute 1 part 10X Collagenase/Hyaluronidase stock with 9 parts complete EpiCult™-C Medium and add to the minced tissue in the dissociation flasks.
Note: Alternatively, mammary tissue can be dissociated in DMEM/F-12 with 15 mM HEPES supplemented with 2% w/v Fraction V BSA to avoid influences of exogenous growth factors and FBS; however, this may result in lower total viable cell yields.
- Ensure that the tissue is well suspended in the enzyme mixture and the final volume is level with the widest portion of the flask. Cover the opening of the flask with sterile aluminum foil.
- Gently dissociate the minced tissue on a rotary shaker at 37°C until all large tissue fragments are digested to liberate ductal and lobular fragments. Typical digestion time is 16 hours (overnight) for normal human mammary tissue. Longer digestion times may be required for tough fibrous tissue; shorter digestion times for softer tissue.
Note: The flasks should be sealed with Parafilm® if the rotary shaker is not in a 5% CO₂ incubator.
- Transfer the dissociated tissue to a 50 mL conical tube, and centrifuge at 80 x g for 30 seconds.
- Discard the overlying liquefied fat layer. The pellet (“A” pellet) is highly enriched for terminal ductal lobular unit (TDLU) epithelial fragments.
- Transfer the supernatant to a new 50 mL conical tube and centrifuge at 200 x g for 3 minutes. The pellet (“B” pellet) from this second centrifugation contains variable numbers of epithelial cells, stromal cells, and red blood cells.
- The supernatant from the second centrifugation is enriched for human mammary fibroblasts. To collect these fibroblasts, transfer the supernatant to a new 50 mL conical tube and centrifuge at 350 x g for 5 minutes.
- The different cell fractions can now be cryopreserved or used in the steps outlined below. For cryopreservation, we recommend that the cells are cryopreserved between 2 million - 10 million cells per cryovial in complete EpiCult™-C Medium (Human) supplemented with 50% FBS and 6% dimethyl sulfoxide (DMSO) or CryoStor® CS10 (Catalog # 07930).
Dissociation of Ductal and Lobular Fragments to Single Cells
- Add 1 - 5 mL of pre-warmed Trypsin-EDTA to pellet “A” from step 7 of the previous section, the Collagenase/Hyaluronidase-dissociated mammary cells, and resuspend.
Note: For the purest epithelial cells, the best starting materials are “A” pellets. “B” pellets may also be used; however, these pellets are more variable in their epithelial cell content.
- Gently pipette up and down with a 1 mL pipettor for 1 - 3 minutes. The sample should become very stringy due to lysis of dead cells and the release of DNA.
- Add 10 mL of cold HBSS with 10 mM HEPES, Without Phenol Red, supplemented with 2% FBS and centrifuge at 350 x g for 5 minutes.
Note: The HBSS + FBS solution is now referred to as HF.
- Remove as much of the supernatant as possible. The cells may be a large “stringy mass” floating in the HF.
- Add 2 mL of pre-warmed 5 U/mL Dispase and 200 μL of 1 mg/mL DNase I Solution. Pipette the sample for 1 minute with a 1 mL pipettor to further dissociate cell clumps. The sample should now be cloudy, but not stringy. If still stringy, add an additional 100 µL of DNase I Solution and pipette as above.
- Dilute the cell suspension with an additional 10 mL of cold HF and filter the cell suspension through a 37 µm Reversible Strainer into a new 50 mL conical tube. Centrifuge at 350 x g for 5 minutes and discard the supernatant.
- If the cell pellet is heavily contaminated with red blood cells, resuspend the pellet in a 1:4 mixture of cold HF:Ammonium Chloride Solution, centrifuge at 450 x g for 5 minutes, and discard the supernatant.
Staining of Single Cell Suspensions with ALDEFLUOR™
- If the breast cells will be stained with antibodies in addition to ALDEFLUOR™ staining, then pre-block the cells with HF supplemented with 10% normal serum from the same species as the primary antibodies (if using directly conjugated antibodies) or from the same species as the secondary antibodies (if using secondary antibodies). Pre-block the cells on ice for 10 min.
- During the pre-block, remove 10 μL of cells from step 1 and add these to 10 μL Trypan Blue and 80 μL HF for counting. Count cells using a hemocytometer.
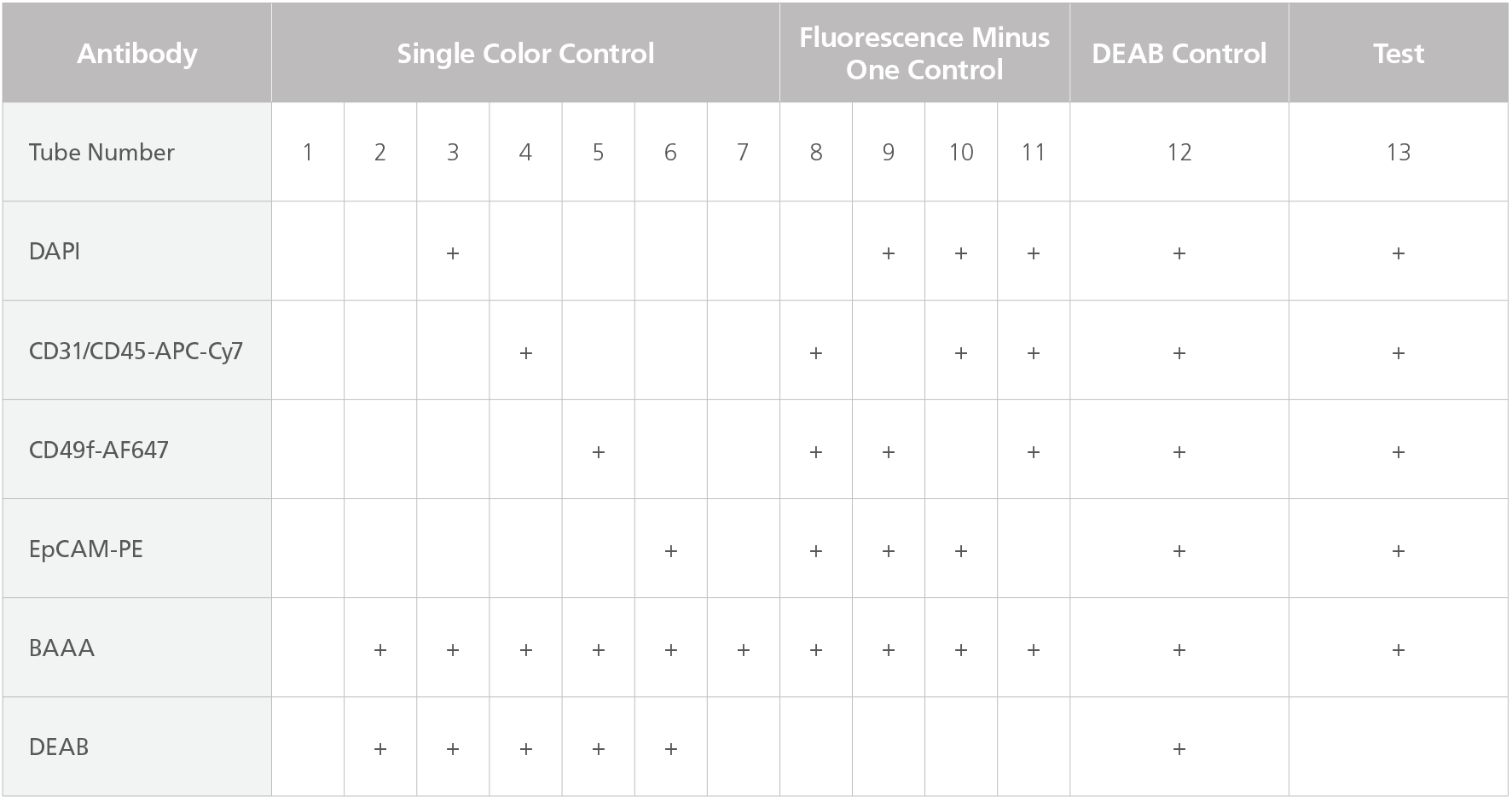
- Label 13 polystyrene 5 ml tubes 1-13. Tube 2 will be the “DEAB control” tube, and tube 7 will be the “BAAA test” tube.
- Remove a small aliquot (~5%) of the pre-blocked cells from step 1 and place these in tube 1. Add some HF to bring the total volume to 500 μL and place these on ice.
- Spin the remaining tubes at 450 × g for 5 min at 4°C. Discard the supernatant.
- Resuspend the cells up to a maximum concentration of 5 × 106 cells/mL in Assay Buffer and then transfer the cells into tube 7.
- For every mL of Assay Buffer in tube 7, add 5 μL of ALDEFLUOR™ DEAB Reagent to the “DEAB control” tube 2. Recap control tube and DEAB vial immediately.
- Add 5 μL of the activated ALDEFLUOR™ reagent per milliliter of sample to tube 7. Mix and immediately transfer half of this mixture to the “DEAB control” tube 2 and mix.
- Incubate tubes 2 and 7 for 30 min at 37 °C.
- Following incubation, evenly distribute the cells that are in tube 2 between tubes 2-6 and tube 12. Similarly, following incubation, evenly distribute the cells that are in tube 7 between tubes 7-11 and tube 13.
- Centrifuge tubes 2-13 at 450 × g for 5 min at 4 °C and discard the supernatant.
- Resuspend the cell pellets in tubes 2, 3, and 7 in 0.5 mL of ALDEFLUOR™ Assay Buffer and place these tubes on ice for the remainder of the staining protocol.
- Stain cells in tubes 4-6 and 8-13 with the primary antibodies outlined in Table 1. The primary antibodies should be diluted in ALDEFLUOR™ Assay Buffer, and incubation should be for a minimum of 10 min on ice.
- After primary antibody incubation, add cold HF media to tubes 3-6 and 8-13 to a final volume of 3 mL, and spin the tubes at 450 × g for 5 min at 4 °C. Discard supernatant, resuspend the cells in tubes 3 and 9-13 in 3 mL of HF supplemented with 1 μg/mL of DAPI, and filter the cells into new round bottom tubes with a cell strainer cap (Catalog #38030). Centrifuge the cells at 450 × g for 5 min at 4°C and resuspend the cells in a small volume (e.g. 200 - 1000 μL) of ALDEFLUOR™ Assay Buffer. The cells are now ready for flow cytometric analysis and sorting.
Note: The SK-BR-3 and MCF-7 breast cancer cell lines can serve as positive and negative controls for ALDEFLUOR™ staining, respectively.
Table 1. Staining Matrix Table
References
Publications Citing the Use of the ALDEFLUOR™ Kit with Different Tissue Types
Mammary
- Simões BM et al. (2015) Anti-estrogen resistance in human breast tumors is driven by JAG1-NOTCH4-dependent cancer stem cell activity. Cell Rep 12(12):1968–77.
- Eirew P et al. (2012) Aldehyde dehydrogenase activity is a biomarker of primitive normal human mammary luminal cells. Stem Cells 30(2): 344–8.
- Shehata M et al. (2012) Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 14(5): R134.
- Marcato P et al. (2011) Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells 29(1): 32–45.
- Ginestier C et al. (2007) ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1(5): 555–67.
- Honeth G et al. (2014) Aldehyde dehydrogenase and estrogen receptor define a hierarchy of cellular differentiation in the normal human mammary epithelium. Breast Cancer Res. 16(3): R52.
Pancreatic
- Visus C et al. (2011) Targeting ALDH (bright) human carcinoma-initiating cells with ALDH1A1-specific CD8⁺ T cells. Clin Cancer Res. 17(19): 6174–84.
- Harbuzariu A et al. (2017) Leptin-Notch signaling axis is involved in pancreatic cancer progression. Oncotarget 8(5): 7740–52.
- Lin L et al. (2016) STAT3 as a potential therapeutic target in ALDH+ and CD44+/CD24+ stem cell-like pancreatic cancer cells. Int J Oncol 49(6): 2265–74.
- Kim-Muller JY et al. (2016) Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic β cells in diabetic mice. Nat Commun 7: 12631.
Colon/Colorectal
- Chen S et al. (2016) Inhibition of PI3K/Akt/mTOR signaling in PI3KR2-overexpressing colon cancer stem cells reduces tumor growth due to apoptosis. Oncotarget 8(31): 50476–88.
- Giraud J et al. (2016) Autocrine secretion of progastrin promotes the survival and self-renewal of colon cancer stem-like cells. Cancer Res 76(12): 3618–28.
- Dalerba P et al. (2007) Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 104(24): 10158–63.
- Huang EH et al. (2009) Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res 69(8): 3382–9.
Neural
- Choi SA et al. (2014) Identification of brain tumour initiating cells using the stem cell marker aldehyde dehydrogenase. Eur J Cancer 50(1): 137–49.
- Ginisty A et al. (2015) Evidence for a subventricular zone neural stem cell phagocytic activity stimulated by the vitamin K-dependent factor protein S. Stem Cells 33(2): 515–25.
- Cheng P et al. (2016) FOXD1-ALDH1A3 signaling is a determinant for the self-renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res. 76(24): 7219–30.
Lung
- Shao C et al. (2014) Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res 20(15): 4154–66.
- Cortes-Dericks L et al. (2014) Cisplatin-resistant cells in malignant pleural mesothelioma cell lines show ALDH(high)CD44(+) phenotype and sphere-forming capacity. BMC Cancer 14: 304.
Ovarian
- Bai S et al. (2016) EGFL6 regulates the asymmetric division, maintenance, and metastasis of ALDH+ ovarian cancer cells. Cancer Res 76(21): 6396–409.
- Silva IA et al. (2011) Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res 71(11): 3991–4001.
- Kryczek I et al. (2012) Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int J Cancer 130(1): 29–39.
Liver
- Chen MH et al. (2016) ALDH1A3, the major aldehyde dehydrogenase isoform in human cholangiocarcinoma cells, affects prognosis and gemcitabine resistance in cholangiocarcinoma patients. Clin Cancer Res 22(16): 4225–35.
- Dollé L et al. (2012) Successful isolation of liver progenitor cells by aldehyde dehydrogenase activity in naïve mice. Hepatology 55(2): 540–52.
Cardiac
- Lee JH et al. (2017) Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell 21(2): 179–94.
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration



