Mechanisms and Applications of Conditional Reprogramming Technology
What is Conditional Reprogramming?
Conditional reprogramming (CR) is a cell culture method in which epithelial cells are seeded onto an irradiated 3T3-J2 mouse fibroblast feeder layer in a specialized medium that contains Y-276321,2, an inhibitor of Rho-associated, coiled-coil containing protein kinase 1 (ROCK1) and ROCK23,4. Epithelial cells, when placed in such culture conditions, are reprogrammed into a highly proliferative undifferentiated adult epithelial stem cell state and are termed conditionally reprogrammed cells (CRCs). These cells form discreet epithelial clusters that are easily distinguishable among the 3T3-J2 feeders in the culture plate (Figure 1). These CRCs can then be grown and passaged indefinitely in adherent culture while maintaining genomic stability.1,2 Cells can be harvested from any passage and seeded into an assay that promotes a specific form of differentiation (e.g. organoid assay, air-liquid interface culture, transplanted into immune-deficient mice), and the cells will differentiate and recapitulate the original tissue morphology, hence the term conditional reprogramming.1,2,5,6 This technology works for all types of epithelia, whether it be a simple, pseudostratified, or stratified epithelium. For example:
- Human cervical cells can generate a stratified epithelium in vitro that resembles the in vivo cervical epithelium, even after >200 population doublings (43 passages) in CR culture.2
- Human bronchial epithelial cells (HBECs), when expanded in CR culture, can still retain the potential to generate histologically normal-looking outgrowths when seeded into air-liquid interface culture.2,7
- Human prostate CRCs can generate normal-looking outgrowths when transplanted with urogenital sinus mesenchyme under the renal capsule.6
CR technology also works across multiple species other than human, including mouse, rat, pig, horse, rabbit, dog, fish, and others.8–14 It has also been reported to work for some non-epithelial cell types, such as endocrine and neuroendocrine cells.6
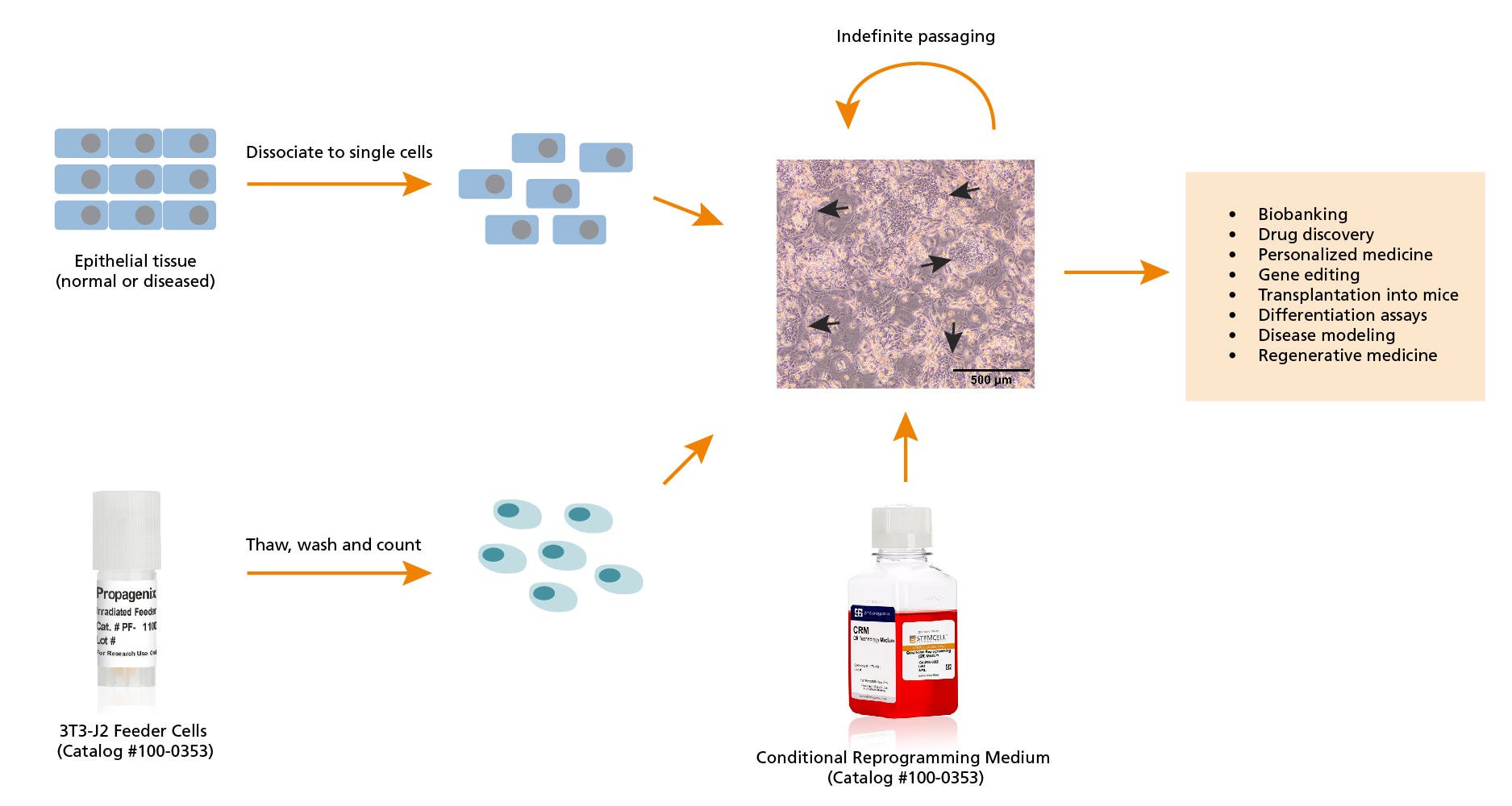
Figure 1. Workflow for Culturing Cells in Conditional Reprogramming Medium
Dissociated epithelial cells are seeded onto irradiated 3T3-J2 feeder cells in Conditional Reprogramming Medium. These conditionally reprogrammed cells (CRCs) will form discrete colonies (arrowheads) in approximately 5 days. The cells between the colonies are the non-proliferative 3T3-J2 feeder cells. Cells can be passaged indefinitely in conditional reprogramming culture and can be used for a variety of downstream applications. Scale bar = 500 µm.
Mechanisms of Conditional Reprogramming Technology
CRCs do not upregulate the pluripotent stem cell transcription factors SOX2, NANOG, or OCT4 to obtain a stem cell state2; instead, multiple other mechanisms appear to be responsible for how CR works. One mechanism is that Δ133p53α, an isoform of p53, is upregulated in CRCs, but not in cells cultured in conventional medium.15 This Δ133p53α isoform inhibits p53 in a dominant negative fashion and thus inhibits p53 effects in promoting senescence and apoptosis.16 The Δ133p53α isoform also upregulates the expression of telomerase.15 Another study indicates that upregulation of poly (ADP-ribose) polymerase 1 (PARP-1), in combination with crosstalk with reactive oxygen species, inhibits G1 cell cycle arrest and results in the extended lifespan of CRCs.17 Other reports have demonstrated that CR conditions increase the amount of activated nuclear β-catenin in CRCs18, and that the irradiated 3T3-J2 feeder cells secrete growth factors such as hepatocyte growth factor19 and secrete extracellular matrix proteins that may support CRC attachment and growth20.
Wallchart: Growing Organoids From Stem Cells
Get an overview of key culture conditions and organoid-forming cells from a variety of epithelial tissues.
Applications of Conditional Reprogramming Technology
The robust expansion of epithelial cells without genetic modification and restoration of tissue-specific progenitor phenotypes is what enables CR technology to complement many applications related to biobanking, disease modeling, drug discovery and development, and regenerative medicine, as outlined below:
- Biobanking: The expansion of rare or small samples of diseased cells permits more experiments to be performed from a single sample, offering opportunities for the biobanking and distribution of cells to the broader research community. In a recent publication, a 100% success rate was reported in culturing 25 human primary prostate tumors.21 With minor changes to the protocol, the technology can be adapted for the expansion of HBECs from Cystic Fibrosis donors7 and to grow colon, pancreatic, and gastrointestinal stromal tumors, as well as other tumor types, with success rates reported between 50-100%5,6,22–26.
- Drug screening for personalized medicine: Cells are expanded very quickly in vitro with a rate typically between 0.5 – 1 population doubling/day, permitting a quick turn-around time for applications such as drug screening.26,27
- Regenerative medicine: Work to date has focused on using CRCs derived from bronchial epithelium for the re-epithelization of the trachea in animal models.28–31 In one study, when HBECs were expanded in Conditional Reprogramming Medium, all cells from both the upper and lower airway of decellularized mouse lungs were reconstituted, including type I and type II alveolar cells.28
Products for Conditional Reprogramming Cell Culture
Related Resources
References
- Liu, X. et al. (2012) ROCK Inhibitor and Feeder Cells Induce the Conditional Reprogramming of Epithelial Cells. Am J Pathol 180(2): 599–607.
- Suprynowicz, F. A. et al. (2012) Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc Natl Acad Sci USA 109(49): 20035–40.
- Ishizaki, T. et al. (2000) Pharmacological properties of Y-27632, a specific inhibitor of rho-associated kinases. Mol Pharmacol 57(5): 976–83.
- Davies, S. P. et al. (2000) Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J 351(Pt 1): 95–105.
- Palechor-Ceron et al. (2019) Conditional Reprogramming for Patient-Derived Cancer Models and Next-Generation Living Biobanks. Cells 8(11): 1327.
- Liu, X. et al. (2017) Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat Protoc 12(2): 439–51.
- Gentzsch, M. et al. (2017) Pharmacological Rescue of Conditionally Reprogrammed Cystic Fibrosis Bronchial Epithelial Cells. Am J Respir Cell Mol Biol 56(5): 568–74.
- Alamri, A. M. et al. (2016) Primary cancer cell culture: mammary-optimized vs conditional reprogramming. Endocr Relat Cancer 23(7): 535–54.
- Kabbesh, H. et al. (2021) Long-term maintenance of viable adult rat sertoli cells able to establish testis barrier components and function in response to androgens. Cells 10(9): 2405.
- Alkhilaiwi, F. et al. (2018) Long-term expansion of primary equine keratinocytes that maintain the ability to differentiate into stratified epidermis. Stem Cell Res Ther 9(1): 181.
- Gardell, A. M. et al. (2014) Derivation and osmotolerance characterization of three immortalized tilapia (oreochromis mossambicus) cell lines. PLoS One 9(5): e95919.
- Saffari, P. S. et al. (2019) Most canine ameloblastomas harbor HRAS mutations, providing a novel large-animal model of RAS-driven cancer. Oncogenesis 8(2): 11.
- Wang, L. et al. (2018) Conditional reprogrammed human limbal epithelial cells represent a novel in vitro cell model for drug responses. Biochem Biophys Res Commun 499(4): 735–42.
- Liu, X. & Mondal, A. M. (2020) Conditional cell reprogramming for modeling host‐virus interactions and human viral diseases. J Med Virol 92(11): 2440–52.
- Mondal, A. M. et al. (2018) Δ133p53α, a natural p53 isoform, contributes to conditional reprogramming and long-term proliferation of primary epithelial cells. Cell Death Dis 9(7): 750.
- Horikawa, I. et al. (2017) Δ133p53 represses p53-inducible senescence genes and enhances the generation of human induced pluripotent stem cells. Cell Death Differ 24(6): 1017–28.
- Liu, L. et al. (2023) Crosstalk of moderate ROS and PARP‐1 contributes to sustainable proliferation of conditionally reprogrammed keratinocytes. J Biochem Mol Toxicol 37(2): e23262.
- Suprynowicz, F. A. et al. (2017) Conditional cell reprogramming involves non-canonical β-catenin activation and mTOR-mediated inactivation of Akt. PLoS One 12(7): e0180897.
- Hynds, R. E. et al. (2018) Cross-talk between human airway epithelial cells and 3T3-J2 feeder cells involves partial activation of human MET by murine HGF. PLoS One 13(5): e0197129.
- Alitalo, K. et al. (1982) Extracellular matrix proteins of human epidermal keratinocytes and feeder 3T3 cells. Journal of Cell Biology 94(3): 497–505.
- Wang, Z. et al. (2023) Integrative multi-omics and drug–response characterization of patient-derived prostate cancer primary cells. Signal Transduct Target Ther 8(1): 175.
- Kettunen, K. et al. (2019) Personalized drug sensitivity screening for bladder cancer using conditionally reprogrammed patient-derived cells. Eur Urol 76(4): 430–34.
- Mimoto, R. et al. (2019) Clinical implications of drug-screening assay for recurrent metastatic hormone receptor-positive, human epidermal receptor 2-negative breast cancer using conditionally reprogrammed cells. Sci Rep 9(1): 13405.
- Chen, C. et al. (2017) A multiplex preclinical model for adenoid cystic carcinoma of the salivary gland identifies regorafenib as a potential therapeutic drug. Sci Rep 7(1): 11410.
- Wu, X. et al. (2020) Conditional reprogramming: next generation cell culture. Acta Pharm Sin B 10(8): 1360–81.
- Li, Y. et al. (2021) Rapid screening for individualized chemotherapy optimization of colorectal cancer: A novel conditional reprogramming technology-based functional diagnostic assay. Transl Oncol 14(1): 100935.
- Yuan, H. et al. (2012) Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N Engl J Med 367(13): 1220–7.
- LaRanger, R. et al. (2018) Reconstituting mouse lungs with conditionally reprogrammed human bronchial epithelial cells. Tissue Eng Part A 24(7-8): 559–68.
- Butler, C. R. et al. (2016) Rapid expansion of human epithelial stem cells suitable for airway tissue engineering. Am J Respir Crit Care Med 194(2): 156–68.
- Hamilton, N. J. I. et al. (2019) Using a three-dimensional collagen matrix to deliver respiratory progenitor cells to decellularized trachea in vivo. Tissue Eng Part C Methods 25(2): 93–102.
- Gowers, K. H. C. et al. (2018) Optimized isolation and expansion of human airway epithelial basal cells from endobronchial biopsy samples. J Tissue Eng Regen Med 12(1) e313–e317.
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration