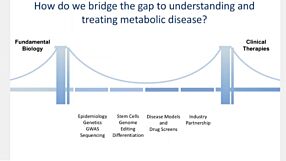
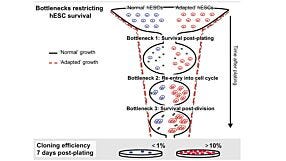
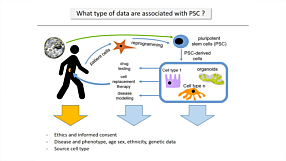
Whole Exome Sequencing Reveals Selective Pressures and Dominant Negative Mutations in hPSC Cultures
View this webinar presentation by Prof. Kevin Eggan (Harvard University) and Dr. Florian Merkle (University of Cambridge) to learn more about genetic variations in human pluripotent stem cell (hPSC) cultures revealed by whole exome sequencing of more than 100 of the world’s hES cell lines, and the importance of maintaining high-quality hPSCs for regenerative medicine and other applications.
The research discussed in the webinar is now published in Nature: Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations.
Publish Date:
April 19, 2017
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration
By submitting this form, you are providing your consent to STEMCELL Technologies Canada Inc. and its subsidiaries and affiliates (“STEMCELL”) to collect and use your information, and send you newsletters and emails in accordance with our privacy policy. Please contact us with any questions that you may have. You can unsubscribe or change your email preferences at any time.