Depletion of Red Blood Cells from Fresh Blood Samples
.
- Document # 29541
- Version 3.0.1
- Mar 2017
Introduction
Red blood cell (RBC) depletion is a critical component of sample preparation for colony-forming unit (CFU) assays of fresh blood. The removal of RBCs minimizes background in assay plates (Figure 1) and ensures that hematopoietic colonies can be counted accurately, whether manually using an inverted microscope or automatically using STEMvision™ (Figure 2). RBCs must be depleted from whole fresh unprocessed blood (i.e. cord blood or peripheral blood), buffy coat fractions of fresh blood, mononuclear cells prepared by density centrifugation (e.g. Lymphoprep™) and blood processed using automated cell separation devices (e.g. Sepax®).
STEMCELL Technologies Inc. has developed a simple protocol that uses HetaSep™ to remove RBCs from fresh blood samples to be tested in the CFU assay. HetaSep™ is an erythrocyte aggregation agent used to quickly separate nucleated cells from RBCs. It’s effectiveness is based on the principle that aggregated erythrocytes settle much faster than dispersed cells. The HetaSep™ protocol requires only 50 μL of blood, takes about 15 - 20 minutes and requires a cell count only at the start of the procedure when cells are to be plated in a CFU assay. The recovery of CFUs in the RBC-depleted fraction is nearly 100% so users can be confident that the HetaSep™ procedure does not adversely affect the number or viability of hematopoietic progenitor cells (Figure 3). These features make it easy to incorporate HetaSep™ into an institution’s CFU assay workflow.
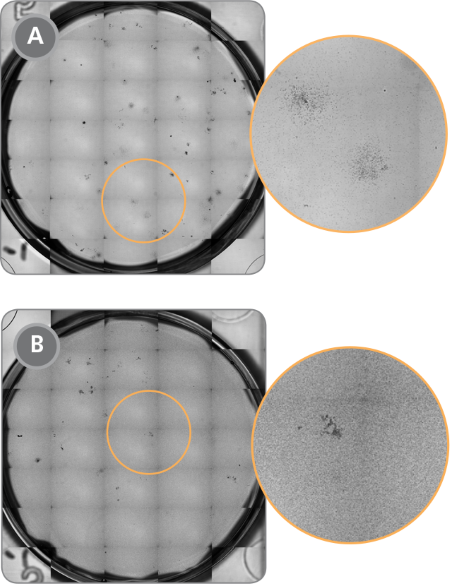
Figure 1. STEMvision™ Images of CFU Assays Plated With and Without Prior Removal of RBCs Using HetaSep™
(A) Acceptable background (minimal RBCs) for CFU assay with prior HetaSep™ treatment and (B) unacceptable background for CFU assay without prior HetaSep™ treatment. Note that fewer colonies are visible due to increased RBC background.
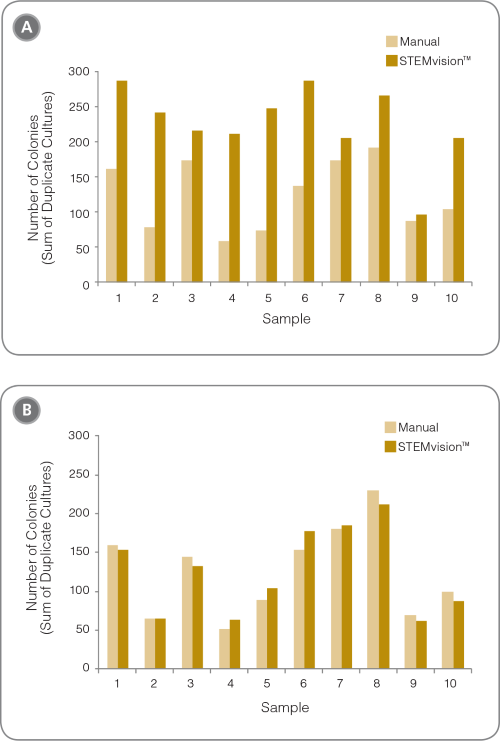
Figure 2. Removal of RBCs is Required for Accurate Counting of CFU Assays With STEMvision™
Shown are the number of colonies that are present in cord blood CFU assays (A) without and (B) with RBC depletion using HetaSep™ before plating (n = 10 fresh whole cord blood samples). The large differences in (A) are due to the high proportion of false positive colonies counted by STEMvision™ in samples with high RBC background.
The recovery of CFUs in the RBC-depleted fraction is nearly 100% so users can be confident that the HetaSepTM procedure does not adversely affect the number or viability of hematopoietic progenitor cells (Figure 3).
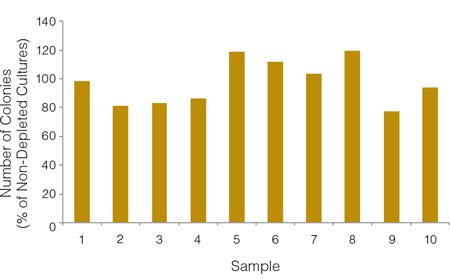
Figure 3. An Average of 97% of Colonies are Recovered Following RBC Depletion With HetaSep™
Cord blood samples (n = 10) were split into two parts, one of which was plated in a CFU assay without RBC depletion and the other having RBCs depleted using the HetaSep™ protocol before plating. Each sample type was plated in duplicate. CFU assays were counted manually and the percent recovery of colonies in each RBC-depleted fraction was calculated relative to results of CFU assays of non-depleted cells from the same donor.
HetaSep™ Protocol for Small-Volume Fresh Blood Samples
- Determine the cell concentration of the initial fresh blood
- Add the following to a 0.5 mL tube:
- 150 μL Dulbecco’s Phosphate Buffered Saline (D-PBS) with 2% fetal bovine serum (FBS) (Catalog #07905)
- 50 μL fresh blood sample
- 40 μL HetaSep™
- Mix thoroughly with a pipette. Avoid creating bubbles. Do not vortex.
- Incubate for 15 - 20 minutes in a 37°C incubator. This step is best done by placing tubes in a culture incubator as opposed to a water bath, as the latter introduces the risk of contamination by residual water when the tube is opened. Note: Cell recovery may be reduced if samples are incubated for more than 20 minutes. If more than 20 minutes has passed, return to step 3 or restart the procedure with fresh sample to ensure optimal recovery.
- Remove the upper phase using a micropipette and transfer to a new tube. Slowly aspirate the upper phase, while moving the tip downward. Do not disturb the RBC pellet (Figure 4B). Note: The size of the RBC pellet may vary due to differences in the hematocrit between sample types. If the RBC pellet is disturbed at any time return to step 3.
- Calculate the cell concentration of the RBC-depleted sample by correcting for the dilution factor of 4.8 (see step 2). For example, if the initial cell concentration was 5 x 106 cells/mL, the concentration of the RBC-depleted sample would be 5 x 106 cells/mL ÷ 4.8 = 1.04 x 106 cells/mL.
- Set up the CFU assay according to the instructions provided in the Technical Manual for Human Colony-Forming Unit (CFU) Assays Using MethoCult™ (Document #28404).
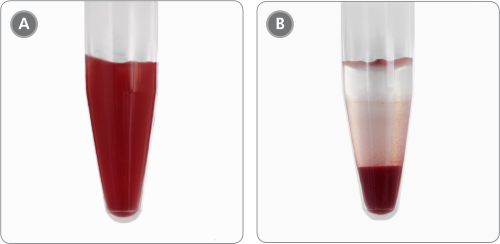
Figure 4. Sample Appearance Before and After Incubation
1.5 mL tube containing cord blood (A) immediately after addition of HetaSep™ and (B) after 20 minute incubation when RBCs have aggregated at the bottom of the tube.
For related products, including specialized culture media and supplements, storage media and antibodies visit www.stemcell.com/CBworkflow or contact us at techsupport@stemcell.com.
Related Resources
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration