Purity Assessment by Flow Cytometry for Lineage-Specific Chimerism Analysis
.
- Document # 29255
- Version 1.1.0
- Nov 2013
The Importance of Purity Assessment in Chimerism Analysis
The EFI and ASHI now officially recognize purity assessment as an essential step in lineage-specific chimerism analysis, yet many chimerism labs are still unfamiliar with this procedure. This Technical Bulletin reviews the importance of lineage-specific chimerism analysis, explains the need for purity assessment, and provides guidelines for assessing purity using flow cytometry.
Isolating Cell Subsets Increases the Sensitivity and Utility of Chimerism Assays
Lineage-specific chimerism analysis is an important tool for monitoring the outcome of allogeneic hematopoietic cell transplantations (allo-HCT). Depending on engraftment success, the host may either achieve full donor chimerism or reach a state of mixed chimerism, in which host hematopoiesis persists alongside donor hematopoiesis in the recipient.1 Chimerism status in transplant recipients can indicate the potential dynamics of disease relapse, graft-versus-host disease (GVHD), (GVT) effects and other outcomes.2-9
Investigating chimerism within specific cell subsets is an increasingly common practice that offers several advantages over analyzing the entire leukocyte population, particularly in recipients of non-myeloablative allo-HCT.10,11 One such advantage is increased assay sensitivity. If a patient has mixed chimerism in only one or a few cell lineages, the overall proportion of host cells may be too low for detection by whole blood assays.1 Furthermore, the significance of mixed chimerism differs between cell subsets. For instance, one recent study found that the degree of mixed chimerism within T and NK cells, but not myeloid cells, allowed patients to be classified into different risk groups for graft rejection.12 In another study, full donor T cell chimerism consistently preceded GVT effects.13 Recent literature suggests that this type of lineagespecific analysis is highly informative for chimerism labs.14-18
Purity Assessment Ensures the Reliability of Lineage-specific Analyses
Performing lineage-specific chimerism analysis can increase the sensitivity of assays and provide essential information. Both of these benefits depend upon the purity of isolated cell subsets, as contamination by non-target cells decreases the reliability of lineage-specific analysis. These issues are particularly common with pathological samples, in which leukocyte numbers and proportions can be variable and atypical.19 Assessing the purity of isolated cell populations is therefore an essential quality control step. As of 2011, the official EFI and ASHI guidelines stipulate that the purity of sorted cell populations must be documented and taken in account when results are analyzed.
When HCE (Haemopoietic Chimaerism and Engraftment) testing is performed on cellular subsets isolated by cell sorting, the purity of the sorted population must be documented and taken into account in the analysis of the results. If this is not possible it must be clearly stated in the report.
- EFI Standards version 5.6.1 (2011), Section I4.190
Document the purity obtained if processing involves isolation of cell subsets. If purity is not assessed, this must be documented on the test report.
- ASHI Standards 2011, Section D.5.3.4.1.8
One of the most common and accurate ways to assess purity is with flow cytometric analysis. The following pages provide step-by-step instructions for performing purity assessments using flow cytometry.
How to Perform Purity Assessments
Purity assessment is critically important in chimerism analysis to ensure that cell subsets are not contaminated by non-target cells. The most common method for purity assessment is flow cytometry, in which target cells are labeled with fluorescent markers and analyzed using a flow cytometer, allowing the proportion of each cell type in the sample to be calculated.
Choosing the Correct Antibodies for Staining
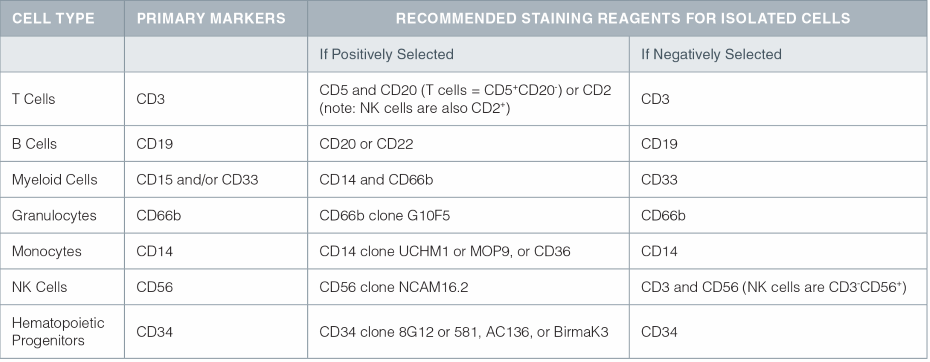
When preparing samples for purity assessment, the appropriate antibodies for staining depend on the cell type and mode of selection (i.e. positive or negative). Table 1 shows the recommended staining reagents for specific cell types.
Some antibodies used in positive selection may fully or partially block the primary cell surface marker. Under these circumstances, there are three different methods of staining for purity assessment:
- Use antibodies targeting alternative cell surface markers on the desired cell population.
- Add a fluorochrome-conjugated antibody (e.g. PE-labeled) targeting the primary cell surface marker at the same time as the selection cocktail.
- Use a fluorochrome-conjugated secondary antibody, such as a FITC-labeled goat anti-mouse IgG. These antibodies will bind to the antibodies used for positive selection.
Negative selection isolates unlabeled target cells. Samples separated by this method can be stained using antibodies against the primary cell surface marker (e.g. CD3 for T cells and CD19 for B cells).
Staining procedures for purity assessment are provided in the Product Information Sheets (PIS) for all EasySep™ and RoboSep™ isolation kits (see product list on final page). Consult the appropriate PIS for detailed instructions.
Table 1. Primary markers and recommended staining reagents for specific cell lineages.
Step-by-Step Protocol: Purity Assessment by Flow Cytometry
Staining Cells for Flow Cytometry
- After cell separation, place 100 µL of enriched cells into two separate 5 mL FACS tubes (cells should be at a concentration between 1 x 106 and 1 x 107 cells/mL).
- To the first tube, add the appropriate fluorescently-conjugated monoclonal antibody or antibodies (see Table 1) according to the antibody manufacturer’s instructions. The volume will typically be 5-20 µL of antibody per test.
- To the second tube, add the appropriate fluorescentlyconjugated isotype control antibody (e.g. FITC mouse IgG1).
- If desired, add a viability stain such as propidium iodide (PI) or 7AAD to each sample. This step allows dead cells to be gated out for more accurate flow cytometry analysis.
- Incubate at 2 – 8®C or on ice for 30 minutes in the dark.
- Wash cells with 1 mL of PBS. Pour off supernatant and resuspend the pellet in 100-500 µL of PBS or FACS sheath fluid. If samples cannot be analyzed immediately, fix with 1% paraformaldehyde and store for up to two weeks at 2 – 8® protected from light.
Gating Sample for Target Cells
In order to obtain an accurate assessment of purity, it is important to gate out cellular debris and dead cells during analysis of the sample. The purpose of purity assessment in chimerism analysis is to ensure that enriched cells are not contaminated by other nucleated cells. Since RBC and debris do not contain nuclei and will not affect downstream DNA-based assays, they should be excluded from the flow cytometry analysis.
To gate the target cell population:
- Create a dot plot of the data displaying FSC vs. SSC, and place a gate around all leukocytes. RBC and debris will be visible in the bottom left corner of the plot, and should be excluded by the gate. (Figure 1, left panel).
- Create a second dot plot of the data displaying FSC vs. the viability stain. Dead cells will be positive for the viability marker and should be excluded by the gate. Figure 1 (right panel) provides an example of gating out dead cells using propidium iodide (PI), a common viability stain.
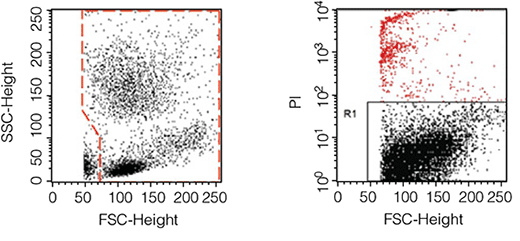
Figure 1. Examples of gates excluding RBC and debris (left) and dead cells (right).
Assessing Sample Purity
Collect 10,000 - 50,000 events for each sample. Sample purity is the percentage of cells positive for the relevant staining antibody in the gated population (Figure 2). The procedure for obtaining purity data will depend on the flow cytometry software used.
In keeping with EFI and ASHI guidelines, document the purity assessment results clearly in your report.
For more details, please contact techsupport@stemcell.com. Guidelines for purity assessment are provided in the Product Information Sheets for all EasySep™ and RoboSep™ kits.
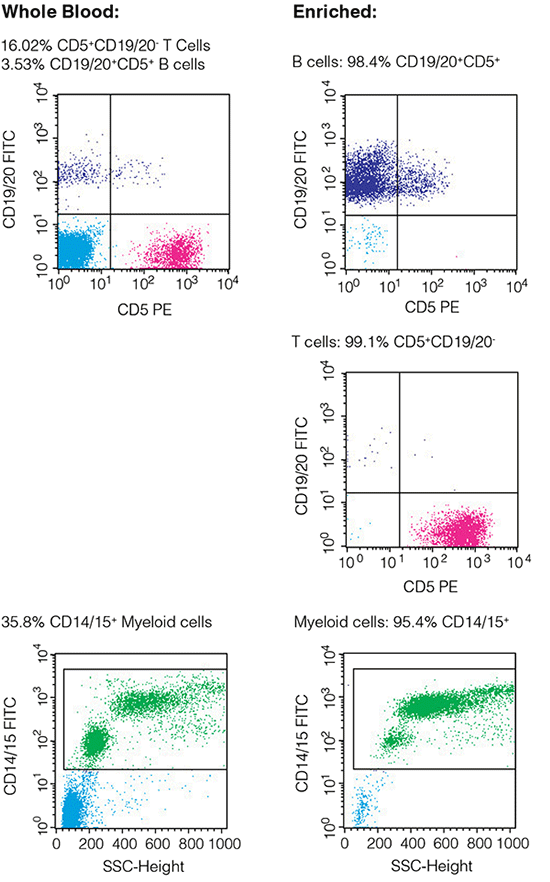
Figure 2. Typical FACS plots before and after enrichment of selected cells (plots show viable (PI-) cells gated on CD45+).
EasySep™ Kits for Isolating Cells for Chimerism Analysis and Recommended Antibodies for Purity Assessment
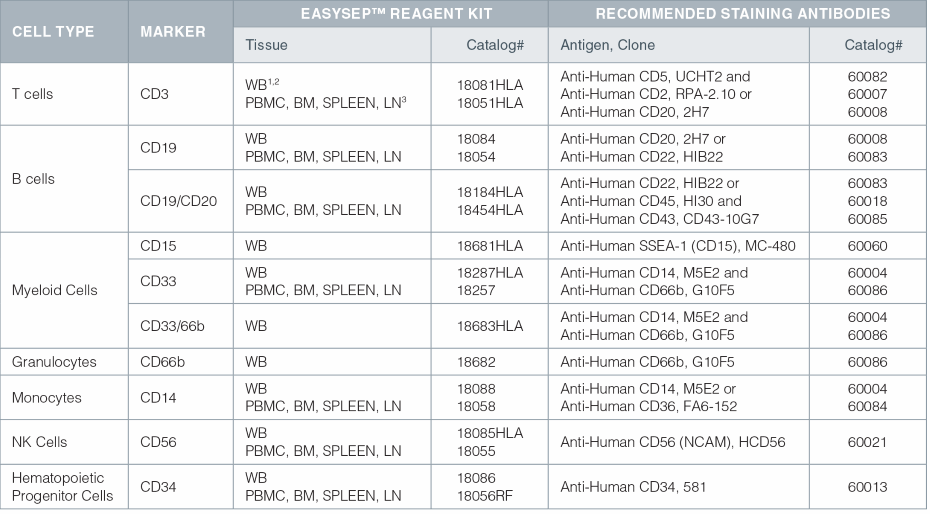
- Anti-Human CD45, HI30 (Catalog# 60018) is also recommended for all whole blood purity assessments.
- WB = Whole Blood; Whole blood kits also work on other red blood cell-containing samples (ie. buffy coat, cord blood, bone marrow).
- PBMC = Peripheral Blood Mononuclear Cell; BM = Bone Marrow; LN = Lymph Node
Antibodies from STEMCELL Technologies
Ensure consistent downstream cell analysis, including purity assessments, by using high-quality primary and secondary antibodies. These antibodies are verified to work with our cell isolation reagents in specific applications.
References
- Antin JH, et al. BBMT. 7:473-485, 2001
- McCann SR, et al. Transfus Apher Sci. 32: 55–61, 2005
- Khan F, et al. Bone Marrow Transplant. 34: 1–12, 2004
- Koldehoff M, et al. Am J Hematol. 81: 735–746, 2006
- Park M, et al. Korean J Hematol. 46: 258-64, 2011
- Perez-Simon JA, et al. Leukemia. 16: 1423–1431, 2002
- Rettinger E, et al. Blood. 118(20): 5681-5688, 2011
- Bader P, et al. Bone Marrow Transplant. 35: 107-119, 2005
- Bader P, et al. Bone Marrow Transplant. 42: S31–S34, 2008
- Childs R, et al. Blood. 94(9): 3234-3241, 1999
- Urbano-Ispizua A, et al. Blood. 97:383-387, 2001
- Breuer S, et al. Leukemia. 26: 509–519, 20012
- Jens G, et al. J Blood Disord Transfus. S1:006, 2012
- Moratto D, et al. Blood. 118: 1675-1684, 2011
- Goh R, et al. Korean J Hematol. 46: 18-23, 2011
- Baron F, et al. Blood. 104: 2254-62, 2004
- Bornhauser M, et al. Haematologica. 94(11): 1613-1617, 2009
- Buno I, et al. Blood. 106(11): 407B, 2005
- Mandigo K, et al. CIM. 18(5): 349-356, 1996
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration


