Endothelial Protein C Receptor (EPCR): A New Marker for Identification and Positive Selection of Mouse Hematopoietic Stem Cells
.
- Document # 28779
- Version 3.0.0
- Jan 2017
Background
Endothelial Protein C Receptor (EPCR, also known as CD201) was originally identified on the surface of endothelial cells and is involved in the regulation of coagulation and inflammation through binding to protein C and activated protein C.1,2 EPCR was initially identified as a hematopoietic stem cell (HSC) marker by gene expression profiling and RT-PCR analysis of highly purified primitive hematopoietic cells that had been isolated using existing HSC markers, such as the c-KIT (CD117) and SCA1 antigens.3-5
Subsequent studies demonstrated that purified EPCR+ mouse bone marrow (BM) cells were highly enriched for in vivo repopulating stem cells.6 Over 90% of primitive BM hematopoietic cells (identified on the basis of their side population (SP) phenotype after staining with the DNA-binding dye (Hoechst 33342) were found to express EPCR, compared to only 1% of cells in the main population (MP).6 EPCR expression was highest in the subset of SP cells with the lowest Hoechst 33342 fluorescence intensity, which is the fraction most enriched for HSCs.6 EPCR+ cells were also highly enriched for cells with the classical Lin-SCA1+c-KIT+ (LSK) CD34- phenotype of hematopoietic stem cells and primitive progenitor cells.
Compared to unfractionated BM, purified EPCRbright cells were approximately 1000-fold enriched for in vivo repopulating stem cells, as identified in competitive transplantation experiments.6 Assuming an HSC frequency of ~1 in 10,000 in unfractionated BM,7 this would translate to an HSC frequency of ~1 in 10 EPCRbright cells. This stem cell frequency is similar to that of SP cells and at least 10-fold higher than that of LSK cells. EPCR expression appeared to be more predictive than Hoechst 33342 exclusion for stem cell activity, since EPCR+ MP cells had significant in vivo repopulating activity, while EPCR- SP cells did not.6
EPCR has been used in combination with other markers to isolate mouse HSCs to high purity from adult mouse BM and fetal liver (FL). In FL, only the EPCR+ subset of LSK cells (~1/3 of LSK cells) was able to reconstitute door type hematopoiesis in recipient mice for at least 4 months, whereas EPCR− LSK cells had little reconstituting ability.8
In BM and FL approximately 50% of EPCR+CD45+CD48−CD150+ cells (E-SLAM phenotype) had long-term multilineage repopulating ability in transplantation experiments in which single purified cells were transplanted into individual mice.9 More recently, 67% of SCA1hi ESLAM cells were shown to have single cell long-term multilineage repopulating ability.10 EPCR protein-activated receptor (PAR-1) signaling, induced locally by activated protein C (PC), is important for maintaining functional HSCs in close association with the hematopoietic microenvironment, while thrombin-induced EPCR/PAR-1 signaling leads to EPCR shedding and recruitment of HSCs into the circulation.11 Results of transplantation studies with EPCR mutant mice that cannot be stimulated with PC indicate that the role of EPCR in maintaining HSCs is at least partially mediated by EPCR expressed on stromal cells.12
Most progenitor cells, capable of forming colonies in methylcellulose-based MethoCult™ medium, express EPCR at intermediate levels, while most mature blood cells do not have detectable EPCR expression.6 This indicates that HSCs gradually lose EPCR expression as they differentiate.
Indeed, typically less than 1% of unfractionated mouse BM cells show detectable staining with anti-EPCR antibody, and most of these cells do not express mature blood cell (lineage) antigens (for example, see Figure 1B). It should be noted however that small subsets of mature monocytes, neutrophils and eosinophils can be positively stained with anti-EPCR antibodies, either due to endogenous EPCR expression on these cells or binding of soluble EPCR from the plasma.13-16 These rare EPCR+ mature cells are much less frequent than mature blood cells that express other commonly used stem cell markers, such as c-KIT and SCA1.
Detection of EPCR Expression
EPCR+ cells can be detected by flow cytometry using conjugated antibodies directed against the cell surface EPCR protein. In the example given in Figure 1, unfractionated BM cells from C57BL/6 mice were stained with anti-EPCR biotin and streptavidin tagged with the fluorochrome phycoerythrin (PE). EPCR+ cells in BM represent an extremely rare cell population (0.4% in this example). Almost all EPCR+ cells express CD45 (Figure 1A), confirming their hematopoietic origin. More than half of the EPCR+ cells are Lin- or c-KIT+ (Figures 1B and 1C). Lin-EPCR+ cells were detected almost exclusively in the c-KIT+ BM subset, and comprised only ~10% of c-KIT+ cells (Figure 1D). Further analysis indicated that the Lin-EPCR+ cells also expressed SCA1 (Figure 2).
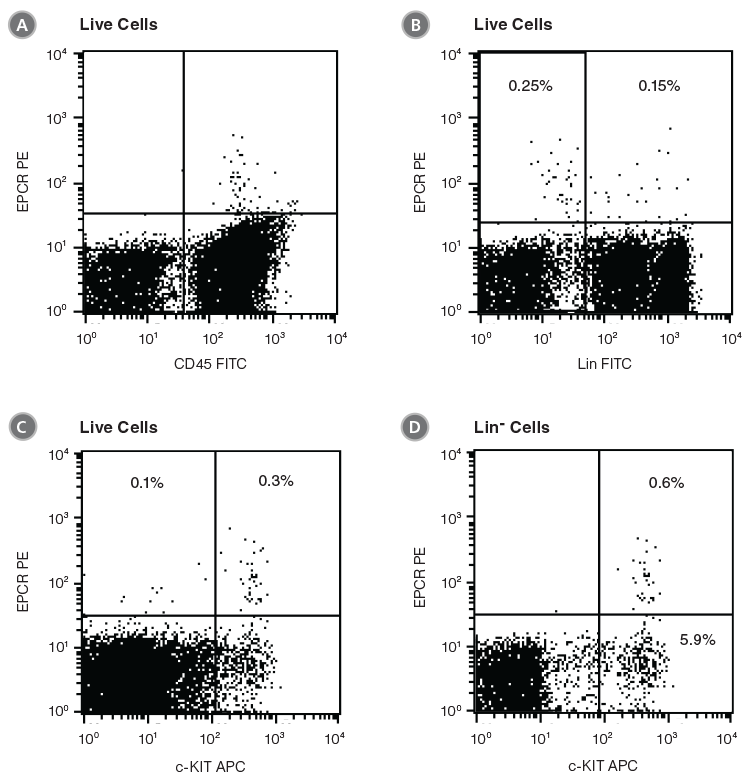
Figure 1. Flow Cytometry Profiles of EPCR Expression on Unfractionated C57BL/6 BM Cells
(A) Cells were stained with anti-EPCR biotin and streptavidin-PE and counterstained with anti-CD45 FITC. (B) Cell staining with a cocktail of FITC-conjugated antibodies directed against myeloid and lymphoid lineage antigens (Lin; CD4, CD8, CD11b, CD11c, CD19, CD45R, Ly-6C/G, NK1.1). (C, D) Cell staining with anti-EPCR biotin/streptavidin-PE and anti-c-KIT APC antibodies. (A), (B) and (C) represent unfractionated BM cells after gating out debris and dead (PI+) cells; (D) represents further cell gating on Lin- cells.
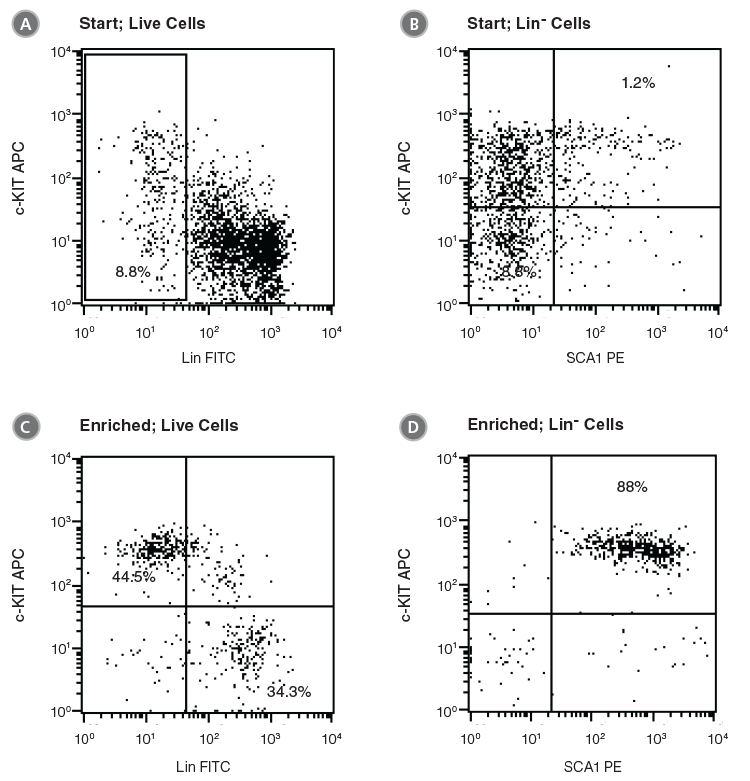
Figure 2. Flow Cytometry Profiles of C57BL/6 BM Cells Before and After EPCR+ Cell Isolation
(A, B) Unfractionated C57BL/6 BM cells and (C, D) EPCR+ cells isolated with the EasySep™ Mouse Biotin Positive Selection Kit (Catalog #18556) were stained with antibodies directed against myeloid and lymphoid lineage antigens (Lin-) in combination with either anti-c-KIT APC or anti-SCA1 PE antibodies. (A) and (C) represent cells after gating out debris and dead (PI+) cells; (B) and (D) represent further gating on Lin- cells.
Isolation of EPCR+ Cells
EPCR+ cells can be rapidly enriched using EasySep™ immunomagnetic cell separation technology. Cells are labeled with biotin- or PE-conjugated anti-EPCR antibody and are bound by bispecific tetrameric antibody complexes (TACs) that recognize the conjugated primary antibody and EasySep™ magnetic particles (Figure 3). Magnetically labeled EPCR+ cells are then retained in the hand-held EasySep™ magnet, while unlabeled cells are simply poured off (Figure 4). EasySep™ magnetic particles are compatible with flow cytometry and other downstream applications, and do not need to be removed following EPCR+ cell isolation.
The optimal method of EPCR+ cell isolation is determined by the proposed downstream usage of the selected cells. It is sometimes desirable for the selected cells to remain unstained with fluorescently labeled antibodies (e.g. to enable three-color flow cytometric analysis with FITC-, PE- and APC-conjugated antibodies). EPCR+ cell isolation using anti-EPCR biotin is recommended in such cases. Accurate measurement of EPCR expression on anti-EPCR biotin-isolated cells is not possible due to blocking of both the EPCR antigen and the biotin molecule with primary antibody and TAC (see Figure 3).
If accurate measurement of EPCR expression on the enriched cells is desired, a PE-conjugated anti-EPCR antibody should be used for cell selection. The PE-labeled EPCR+ cells will be available for flow cytometric analysis immediately after cell separation. The expression of other cell surface proteins can be further analyzed by using appropriately conjugated antibodies (e.g. anti-Lin FITC, anti-SCA1 APC and anti-c-KIT PE-Cy5 for LSK cells).
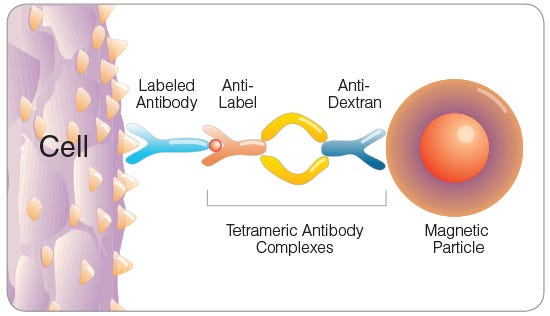
Figure 3. Tetrameric Antibody Complex (TAC) Crosslinking a Cell to a Dextran-Coated Magnetic Particle
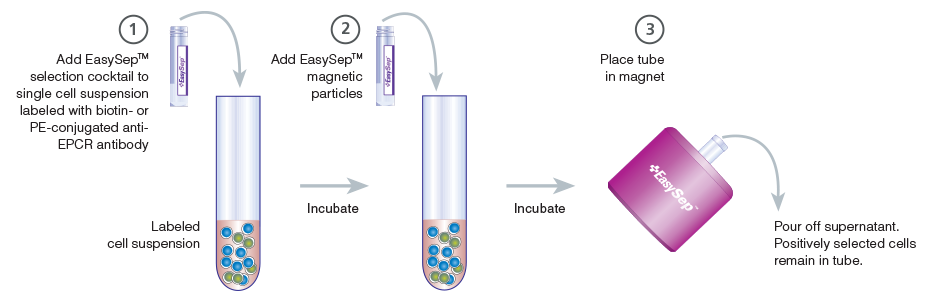
Figure 4. Schematic Drawing Showing EasySep™ Cell Isolation Protocol
EPCR+ cells are targeted with anti-EPCR antibodies and are then crosslinked to magnetic particles. The sample is placed in the EasySep™ Magnet, and the magnetically labeled EPCR+ cells remain in the tube when the supernatant is removed. The EPCR+ cells are easily collected by removing the tube from the magnet.
Table 1. Products for Detecting and Selecting EPCR+ Cells
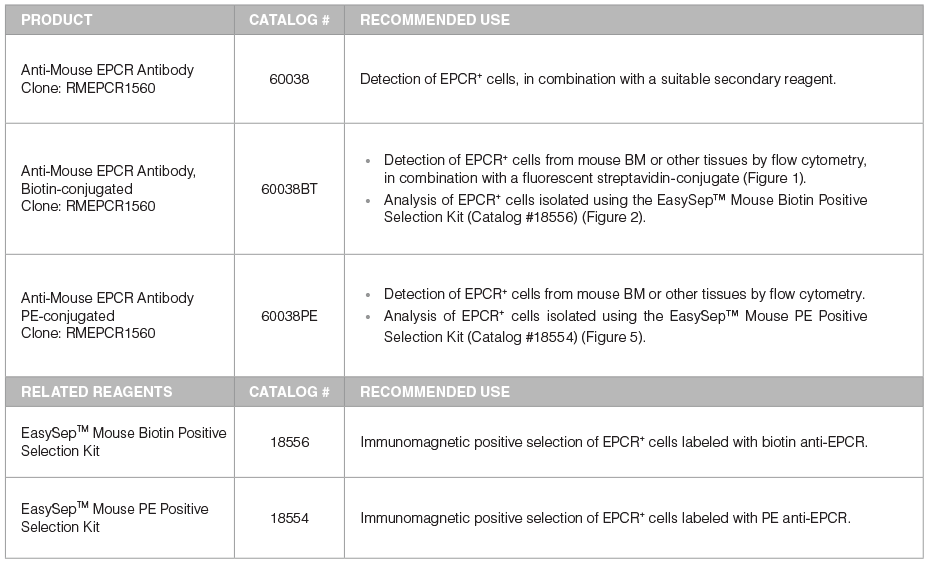
*For more antibodies, including lineage markers, see below.
Example 1: EPCR+ Cell Isolation Using a Biotinylated Anti-EPCR Antibody
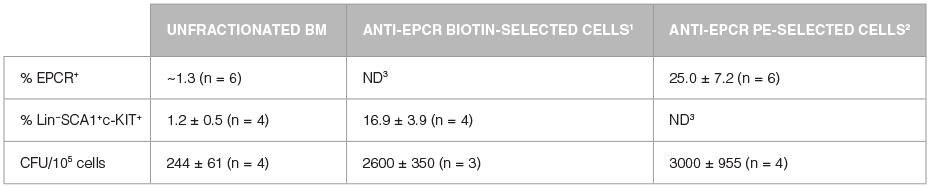
Unfractionated BM cells (Figure 2A,B) and EasySep™-selected EPCR+ cells (Figures 2C, D) were analyzed for lineage antigen (Lin), SCA1 and c-KIT expression. EPCR-selected cells were highly enriched for Lin-SCA1+c-KIT+ cells, whereas Lin+ and Lin-SCA1−c-KIT+ cells were generally not recovered (Figures 2C, D). Enrichment of EPCR+ cells increased the frequency of Lin-SCA1+c-KIT+ cells 15-fold to 16.9 ± 3.9% from a starting frequency of 1.2 ± 0.5% (n = 4). The frequency of hematopoietic colonies, as identified in colony-forming unit (CFU) assays using MethoCult™ GF M3434 medium (Catalog #03434), increased 11-fold compared to unfractionated BM, from 244 ± 61 to 2600 ± 350 CFU/105 plated cells (Table 2, n = 3).
Example 2: EPCR+ Cell Isolation Using a PE-Conjugated Anti-EPCR Antibody
As isolated cells are already labeled with anti-EPCR PE, this approach enables direct analysis of EPCR+ cell purity and recovery after enrichment. The EPCR+ selected cells were stained with anti-Lin FITC and anti-c-KIT APC. Magnetic separation increased the frequency of EPCR+ cells over 20-fold compared to unfractionated BM, from approximately 1% to 25.0 ± 7.2% (Figure 5A,B; Table 2, n = 6). The frequency of CFUs increased 12-fold from 244 ± 61 CFU/105 plated cells to 3000 ± 955 CFU/105 anti-EPCR PE-enriched cells (Table 2, n = 4). Gating on EPCR+ cells revealed that ~26% of EPCR+ cells were Lin- and c-KIT+ (Figure 5D). As demonstrated above (Figure 2), the majority of the Lin-EPCR+c-KIT+ cells are SCA1+ and probably represent very immature progenitors, including HSCs. These Lin-EPCR+c-KIT+SCA1+ cells represent only a small subset of Lin-c-KIT+ cells; most of the Lin-c-KIT+ cells are EPCR−SCA1− and represent more differentiated cells. Approximately 50% of the EPCR+ cells were Lin+ and c-KIT- (Figure 5D), which is consistent with the expression of EPCR on small numbers of mature cells.13-16 Although these Lin+EPCR+ mature cells are relatively abundant compared to the primitive Lin-EPCR+c-KIT+ cells, they represent less than 1% of lineage+ cells in unfractionated BM.
Table 2. Enrichment of Hematopoietic Cells by EasySep™ Positive Selection of EPCR+ Cells Using Biotinylated or PE-Conjugated Anti-EPCR Antibodies
- This approach is recommended if the selected cells are to remain unstained with fluorescently labeled antibodies, e.g. to enable three-color flow cytometry analysis with FITC-, PE- and APC-conjugated antibodies. Accurate measurement of EPCR expression on anti-EPCR biotin selected cells is not possible due to blocking of both the EPCR antigen and the biotin molecule with primary antibody and TAC.
- This approach is recommended if accurate measurement of EPCR expression on the enriched cells is desired. The expression of other cell surface proteins can be further analyzed by using appropriately conjugated antibodies (e.g. anti-Lin FITC, anti-SCA1 APC and anti-c-KIT PE-Cyanine5 for LSK cells).
- ND: Not Determined
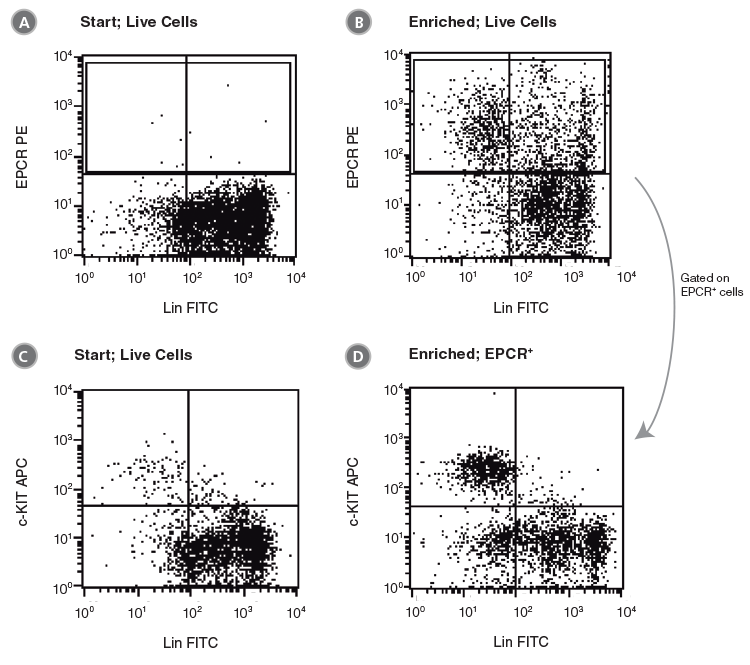
Figure 5. Flow Cytometry Profiles of C57BL/6 BM Cells Before and After EPCR+ Cell Isolation Using Anti-EPCR PE and The EasySep™ PE Selection Kit for Mouse Cells
(A, C) Unfractionated C57BL/6 BM cells and (B, D) EPCR+-selected cells were stained with anti-lineage FITC antibodies in combination with either anti-EPCR PE or anti-c-KIT APC antibodies. (A), (B) and (C) represent cells after gating out debris and dead (PI+) cells; (D) represents further gating on the EPCR+ cell population.
Conclusion
EPCR expression may be used as an alternative to, or in combination with, other markers, such as the LSK, SP or SLAM receptor phenotypes, to identify and isolate mouse HSCs and primitive progenitors.6 Unlike SCA1, EPCR expression can be used to detect and isolate HSCs from BALB/c mice as well as C57BL/6 mice. However, EPCR expression on hematopoietic cells from other mouse strains remains to be demonstrated. Isolation of mouse HSCs on the basis of SP phenotype requires staining with Hoechst 33342 dye and exposure to UV laser light, which is toxic to cells and technically challenging to use. Staining with anti-EPCR antibodies is much more straightforward than Hoechst 33342 staining, does not require a flow cytometer or cell sorter equipped with a UV laser, and does not appear to be harmful to the cells. In addition, it allows rapid pre-enrichment of HSCs and primitive progenitors by immunomagnetic separation. Due to the very low frequency of EPCR+ cells in unfractionated BM, the use of pre-enrichment strategies prior to flow cytometric sorting is highly recommended. This approach will reduce sorting time significantly and may result in better cell purities and recoveries. The recommended pre-enrichment strategies include EasySep™ positive selection of EPCR+ cells (Table 1), SCA1+ cells (Catalog #18756) or c-KIT+ cells (Catalog #18757), or depletion of lineage-positive cells (Catalog #19756).
Finally, gene expression analyses of purified human CD34+CD38-Lin- cells have indicated that EPCR is also expressed on primitive human hematopoietic cells.17 In addition EPCR expression was observed to increase in human CD34+ cells during stimulation with cytokines and the small molecule, UM171, known to promote HSC self renewal.18 These results suggest that EPCR may have a role in human hematopoiesis as well.
See all related products for mouse HSPC research, including specialized culture media, antibodies and cytokines.
References
- Esmon CT (2006) Curr Opin Hematol 13: 382-385.
- Fukudome K et al. (1994) J Biol Chem 269: 26486-91.
- Akashi K et al. (2003) Blood 101: 383-390.
- Ivanova NB et al. (2002) Science 298: 601-604.
- Ramalho-Santos M et al. (2002) Science 298: 597-600
- Balazs A et al. (2006) Blood 107: 2317-2321
- Boggs DR et al. (1982) J Clin Invest 70: 242-253.
- Iwasaki H et al. (2010) Blood 116: 544-553.
- Kent DG et al. (2009) Blood 113: 6342-6350.
- Wilson NK et al. (2015) Cell Stem Cell 16: 712–24.
- Gur-Cohen S et al. (2015) Nat Med 21: 1307-17.
- Pepler L et al. Blood (2015) 126: 673-82.
- Galligan L et al. (2001) Br J Haematol 115: 408-414.
- Feistritzer C et al. (2003) J Allergy Clin Immunol 112: 375- 381.
- Sturn DH et al. (2003) Blood 102: 1499-1505.
- Kurosawa S et al. (2000) J Immunol 165: 4697-4703.
- Georgantas RW 3rd et al. (2004) Cancer Res 64: 4434-4441.
- Fares I et al. (2014) Science, 345: 1509-12.
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration