Identification of Viable Stem and Progenitor Cells with ALDEFLUOR™
.
- Document # 28728
- Version 2.0.0
- May 2023
Introduction
The ALDEFLUOR™ reagent system offers a non-immunological method to identify human stem and progenitor cells on the basis of activity of the enzyme aldehyde dehydrogenase (ALDH). ALDEFLUOR™ was originally developed for detection of hematopoietic stem and progenitor cells (HSPCs) in human bone marrow (BM), mobilized peripheral blood (MPB) and umbilical cord blood (CB), but since then has increasingly been used to identify and characterize stem and progenitor cells in other tissues and to study cancer stem cells in primary tumors and cell lines.
Assay Principle
ALDEFLUOR™ is supplied in the form of Bodipy™-aminoacetaldehyde diethyl acetal (BAAA-DA) (Figure 1), which, by itself, is not a substrate of ALDH. BAAA-DA is dissolved in dimethylsulfoxide (DMSO) and exposed to acid to convert it into Bodipy™-aminoacetaldehyde (BAAA), which is a fluorescent substrate for ALDH. BAAA is not stable for long periods at room temperature, but can be stored for several months when frozen in aliquots. Treatment of cells is usually performed at 37°C with BAAA at a concentration of 1.5 μM. BAAA is uncharged and can diffuse freely across the plasma membrane of intact viable cells. Specific intracellular ALDH isoforms convert BAAA into Bodipy™-aminoacetate (BAA), which is retained intracellularly because of its net negative charge, which prevents free diffusion. The assay buffer (supplied with the ALDEFLUOR™ kit) contains a transport inhibitor that prevents efflux of the BAA from the cells. As a result, cells expressing high levels of certain ALDH isoforms retain BAA and thus fluoresce more intensely than cells that don’t express these ALDH isoforms. BAA is only retained in cells with intact plasma membranes, not in apoptotic and dead cells, even if these have high ALDH activity. This makes ALDEFLUOR™ very specific for detecting only viable and metabolically active cells.
Incubation of cells with BAAA in the presence of a 10-fold molar excess of the ALDH inhibitor diethylamino-benzaldehyde (DEAB) results in a significant decrease in the fluorescence intensity of ALDEFLUOR™ bright (ALDHbr) cells and is used as a negative staining control to set up the flow cytometer and to distinguish between ALDHbr cells and more dimly stained (ALDHlow) cells that take up BAAA passively and/or have low levels of ALDH activity.
The fluorescence of ALDEFLUOR™-treated cells is detected in the same fluorescence channel of standard flow cytometers as used for other dyes that emit green fluorescence, e.g., fluorescein. However, the ALDEFLUOR™ fluorescence is usually much brighter than obtained in standard flow cytometry staining experiments with fluorochrome-labeled antibodies. For this reason, the amplification of the green fluorescence detector of the flow cytometer needs to be adjusted to much lower levels than for the detection of fluorescent antibody-stained cells.
ALDEFLUOR™-treated cells can be stained with fluorescently conjugated antibodies against cell surface markers and analyzed by multiparameter flow cytometry to correlate ALDH expression with other phenotypic properties of the cells. An example of this is the ALDHbr Assay Kit (Catalog #01711), which is intended for the simultaneous detection of ALDH activity and CD34 expression on viable CD45+ glycophorin A (GlyA) cells in human CB. This kit contains ALDEFLUOR™, APC-anti-CD34, PE-anti-CD45, PE-Cyanine5-anti-Glycophorin A/B, and 7-AAD viability dye and requires a flow cytometer equipped with 488 nm and 635 nm lasers for excitation and appropriate filters for detecting Bodipy™, APC, PE, PE-Cyanine5 and 7-AAD fluorescence.
For a detailed description of the experimental procedure for ALDEFLUOR™ treatment of hematopoietic cells and detection of ALDHbr cells, please refer to the ALDEFLUOR™ and ALDHbr Assay Kit product information sheets.
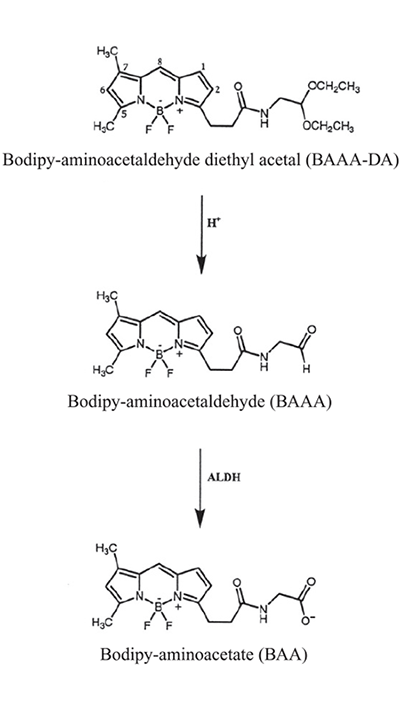
Figure 1. Structure of BAAA-DA, BAAA and BAA
Prior to use, BAAA-DA is dissolved in DMSO and converted to BAAA when exposed to acid (1 N HCl) for 15-30 minutes at room temperature. BAAA can freely diffuse into viable cells and is converted by ALDH into BAA, which is negatively charged and retained inside the cells.
ALDH Isoform Specificity of ALDEFLUOR™
There are 19 ALDH genes in the human genome1; this variety of isoforms ensures that there are many different enzymes available to catalyze a wide variety of different aldehyde substrates. The ALDEFLUOR™ substrate BAAA was initially perceived to be specific for ALDH1A12, since this is the predominant isoform present in human hematopoietic stem cells (discussed below), which was the first application of this assay. However, subsequent studies have demonstrated that the ALDEFLUOR™ substrate BAAA interacts with multiple additional ALDH isoforms, including ALDH1A2, 1A3, 2, 5A1, 6A1, and 7A1, albeit with varying specificities.3–5 ALDEFLUOR™ is not a substrate for ALDH3A1.6
Properties of ALDHbr Hematopoietic Cells
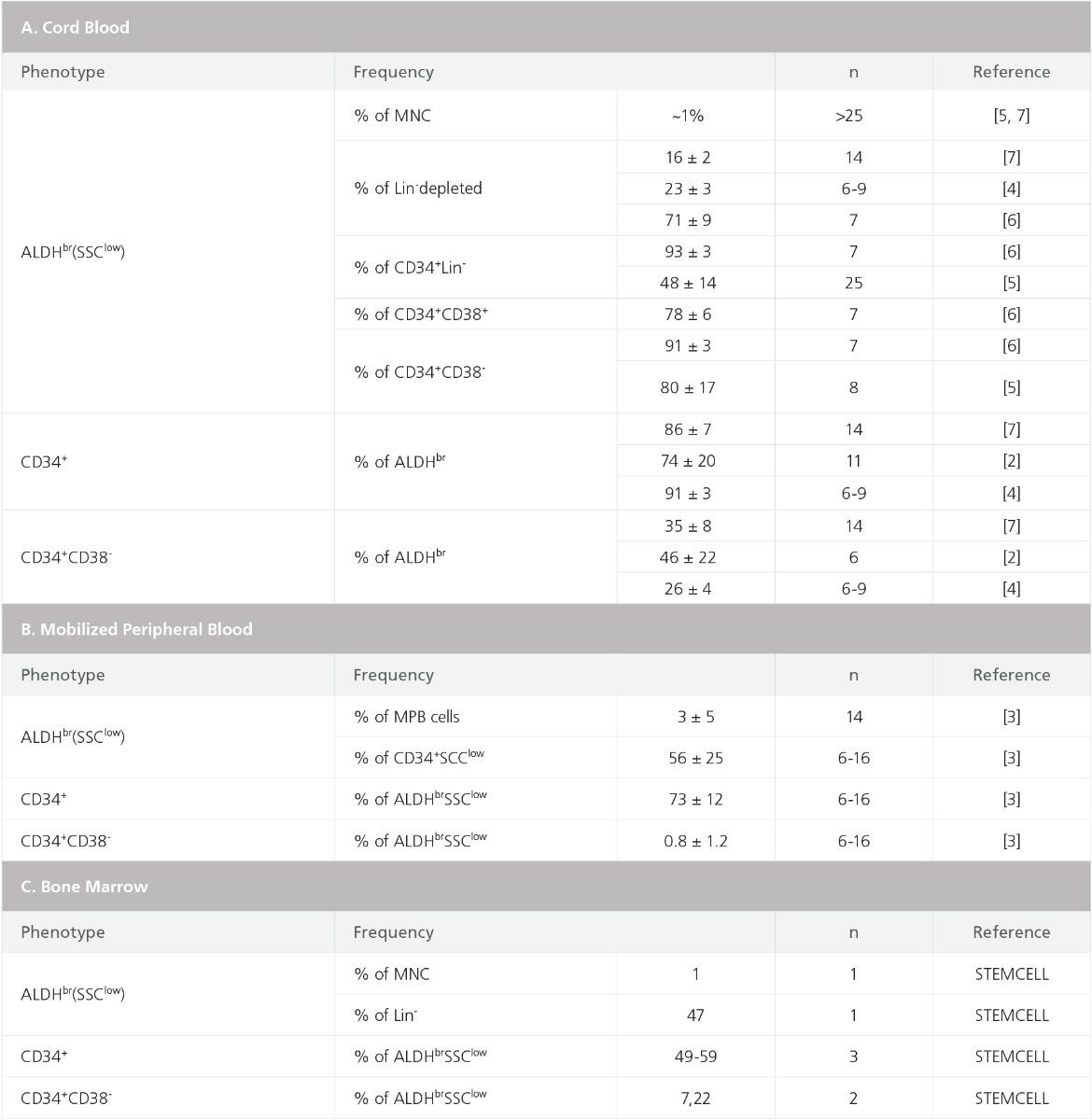
Most ALDHbr cells in BM, MPB, and CB have low side scatter, high expression of HSPC markers (specifically CD34), and absent or low expression of lineage antigens that are expressed on mature cells (Figures 2, 3, and Table 1).7–13 The frequencies of ALDHbr SSClow cells that express CD34 in MPB and CB typically range between 70 and 95%, with somewhat lower percentages for BM, ~50 - 60% (Figure 3C-F and Table 1).10,13 Purified ALDHbr cells, isolated by fluorescence-activated cell sorting, are enriched for NOD/SCID repopulating cells as well as for progenitor cells for all blood cell lineages detectable in hematopoietic culture assays, specifically long-term culture initiating cells (LTC-IC) and colony-forming unit (CFU) assays.8,10,13
ALDH activity can be used to measure MPB and CB graft quality and predict hematopoietic recovery after transplantation of human patients.9,14–17 The number of transplanted ALDHbr SSClow cells in autologous MPB stem cell grafts has been shown to correlate with the time to neutrophil and platelet reconstitution.9,14 Similarly, the ALDHbr cell content of thawed CB units, but not the CD34+ content, has been shown to correlate with the CFU-GM content of these cells.16 These results indicate that enumeration of ALDHbr cells may provide an alternative to measurements of CD34+ cell content to identify and enumerate intact, viable hematopoietic stem and progenitor cells in clinical grafts. Such an assay is also much faster than the CFU-assay, which is the recommended potency assay for measuring graft quality.
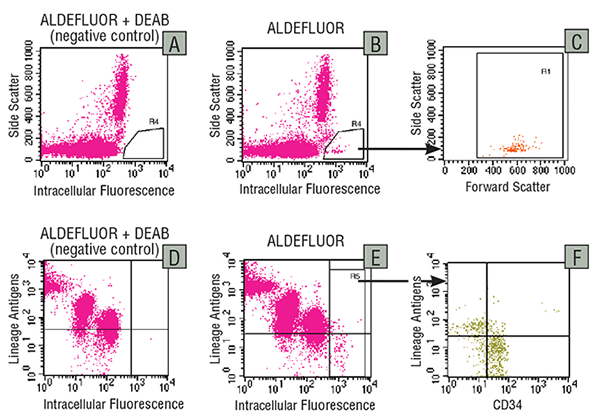
Figure 2. Detection of ALDH expression in human cord blood cells
Human low-density UCB cells were treated with ALDEFLUOR™ in the presence of the ALDH inhibitor DEAB (A,D) or with ALDEFLUOR™ alone (B,C,E,F). A fraction of the cells were analyzed by flow cytometry without further treatments (A-C), whereas a second fraction was stained with a panel of PE-labeled antibodies against lineage-specific antigens (CD2, CD3, CD11b, CD14, CD16, CD19, CD24, CD56 and Glycophorin-A) and Cy-5 labeled anti-CD34 (D-F). A and B show the intracellular fluorescence versus the Side Scatter of cells gated in an electronic window to exclude debris and dead, propidium iodide-stained cells. Region R4 in B shows a brightly fluorescent ALDHbr population (0.3% of cells) that is absent from the DEAB control in A. The Forward versus Side Scatter profile of the ALDHbr cells gated in R4 is shown in C. D and E show the intracellular fluorescence versus lineage antigen expression for all viable cells. F shows CD34 expression versus lineage antigen expression for gated ALDHbr cells (Region R5 in E).
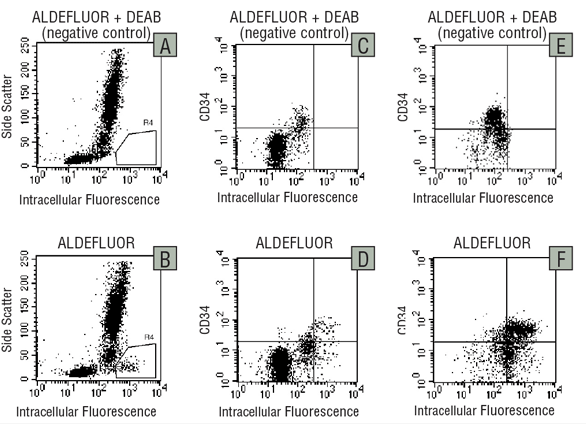
Figure 3. Detection of ALDH expression in human bone marrow cells
Low-density BM cells (A-D) and lineage-depleted* BM cells (E,F) were treated with ALDEFLUOR™ in the presence (A,C,E) or absence (B,D,F) of the ALDH-inhibitor DEAB, and immunostained with Cy-5-conjugated anti-CD34.
*Depletion of cells expressing lineage antigens expressed on mature blood cells can be achieved by immunomagnetic cell separation (e.g. EasySep™ catalog #19056 or 19356) or density centrifugation-based methods (RosetteSep, catalog # 15026, 15027, 15126).
Table 1. Frequencies of ALDH-Expressing Subsets in Human Cord Blood, Mobilized Peripheral Blood, and Bone Marrow
ALDH Expression by Human Leukemia Cells
ALDH activity has also been explored as a marker for malignant cells in myeloproliferative disorders. ALDEFLUOR™ staining patterns of CD34+ cells from patients with the chronic phase of chronic myeloid leukemia (CML) are similar to those of normal CD34+ cells, whereas those from blast crisis CML cells are more heterogeneous.18,19 Similar to their normal counterparts, CD34+CD38- ALDHbr CML cells are enriched for cells that can engraft NOD/SCID mice, whereas ALDHlow cells do not engraft.18
ALDH expression and activity in acute myeloid leukemia (AML) is highly variable between patients, with the proportion of ALDHbr cells ranging from undetectable to over 50%.12,20–22 Most ALDHbr AML cells co-express CD34 and the frequency of ALDHbr cells has been found to correlate significantly with adverse cytogenetic factors and poor prognosis21,22; however, at least one study has reported that there was no association between ALDH patterns, prognosis, or morphological classification.12
Purified ALDHbr cells from patients with a high proportion of ALDHbr cells have leukemic stem cell ability as assessed in NOD/SCID mouse transplantation assays, whereas those from patients with very few (typically ~0.1%) ALDHbr cells mostly consist of normal HSPCs.12,20,21 Two studies have reported that ALDH activity is lower in AML CD34+CD38- cells than on normal CD34+CD38- HSPCs in AML BM, suggesting that differences in ALDH activity may be useful to distinguish normal HSPCs from malignant cells and to detect minimal residual disease after treatment.23,24 However, leukemia-initiating cells are not exclusively CD34+CD38- and can also be CD34+CD38+ or CD34-.Results of other studies indicated that leukemia-initiating cells can have similar high ALDH activity to normal HSPCs.12,20,21 These differences may reflect the large degree of heterogeneity among AML cases and between different AML subclones in individual AML patients. They may also reflect differences in flow cytometry gating approaches used in various studies to identify AML subsets on the basis of ALDH activity and cell surface markers.
Detection of ALDH Activity in Mouse Hematopoietic Stem Cells and Progenitor Cells
The utility of ALDEFLUOR™ for analysis of hematopoietic cells from mice is less obvious than its utility for human HSPCs. High levels of ALDH activity have been identified in mouse hematopoietic cells in experiments that used a different ALDH substrate, dansyl aminoacetaldehyde (DAAA), which can only be detected using flow cytometers equipped with a UV laser.7 In this study a rare population of small ALDHbr cells was identified that was capable of long-term multi-lineage repopulation after transplantation but lacked radioprotection and spleen colony-forming ability.7 Of interest, these cells expressed low to undetectable levels of Thy-1, Sca-1, c-Kit, and CD34, and may represent a different class of stem cells than the cells that express the “classical” Lin- Sca1+Kit+ (LSK) CD34- phenotype. Several other studies using ALDEFLUOR™ confirmed that high ALDH activity is not detectable in most mouse LSK cells or in cells identified with other well-established HSC markers, such as high efflux of Hoechst dyes (“side population”) or CD48-CD150+EPCR+ (ESLAM) phenotypes.25–27 Some HSCs in mouse BM have intermediate ALDH activity; these cells appear to be enriched for cells that are in the G1 phase of the cell cycle and can engraft recipient mice after intra-bone injection, but not after intravenous injection.28 Exposure of mice to the ALDH inhibitor DEAB or ALDH1A1-specific siRNA constructs promotes expansion of HSPCs with radioprotective ability, a property typically attributed to short-term repopulating cells; but hematopoiesis in ALDH1A1 knockout mice is normal and ALDEFLUOR™ fluorescence of unfractionated BM cells and purified HSPCs was indistinguishable to that in wild-type mice.29 These data suggest that although ALDH is expressed in mouse HSPCs, its activity cannot be easily detected using the regular ALDEFLUOR™ system.
Expression of ALDH by Non-Hematopoietic Tissues
ALDHs are widely expressed in non-hematopoietic cells, with one of the best studied being the mammary epithelium. The non-pregnant mammary gland contains three lineages of epithelial cells: estrogen receptor (ER)-expressing, milk lineage, and basal/myoepithelial. The milk lineage cells are a subpopulation of ER- cells present in the gland that will proliferate during pregnancy to generate milk-secreting alveolar cells during lactation.30–32 Experiments in mice demonstrate that each of these lineages is maintained by their own subpopulation of stem cells.33–36 The ALDEFLUOR™ assay identifies a subset of milk lineage stem-like cells (Figure 4) that are highly clonogenic in vitro32,37, have stem cell properties38, and are perceived to be the precursors for basal-like breast cancer39,40. The ALDEFLUOR™ assay also identifies a large proportion of stromal cells within the mammary gland, so additional markers such as EpCAM are required to discriminate ALDHbr epithelial cells from ALDHbr stromal cells.
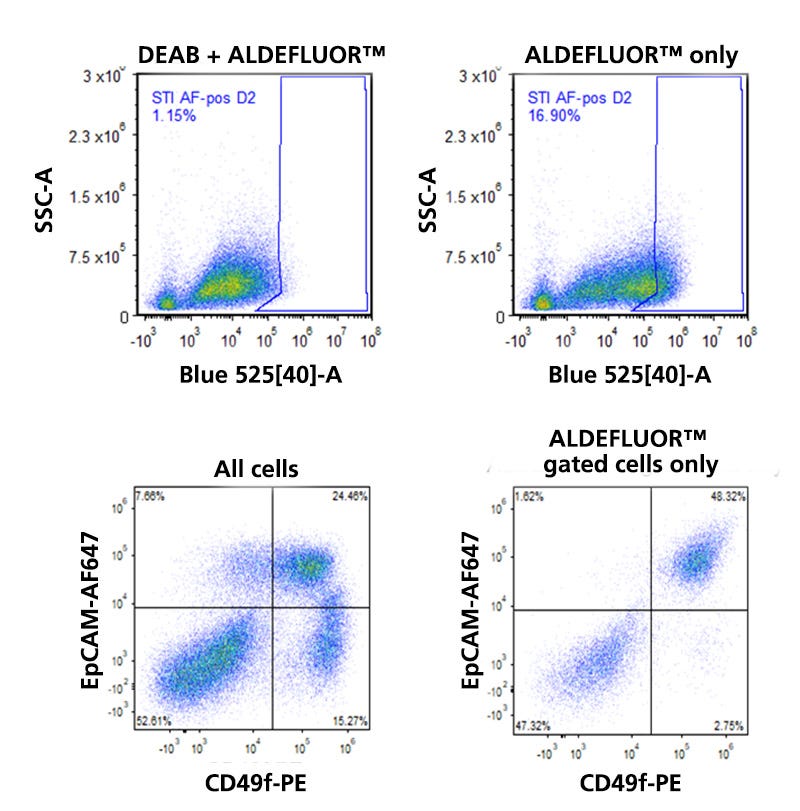
Figure 4. Detection of ALDH Expression in Human Breast Tissue
Normal human breast tissue from a reduction mammoplasty specimen was dissociated and incubated in the presence of Bodipy™ aminoacetaldehyde (BAAA) with or without the ALDH inhibitor diethylamino-benzaldehyde (DEAB). As illustrated in the flow cytometry dot plots in the top row, the presence of DEAB greatly reduces the frequency of ALDHbr events. The bottom left dot plot shows the distribution of EpCAM and CD49f-expressing cells among all viable CD45- cells. The three main mammary lineages, which are estrogen receptor-expressing (EpCAM+CD49f-), milk (EpCAM>+CD49f+), and basal (EpCAMlowCD49f+), are resolvable. Most stromal cells have an EpCAM-CD49f- phenotype. When the mammary cells are gated solely on ALDHbr events, it becomes apparent that high levels of ALDH activity are largely restricted to only a subset of milk lineage and stromal cells.
In some breast tumors, the ALDHbr subpopulation is enriched for cells that can generate tumors when transplanted into immune-deficient recipient mice.38 The presence of ALDHbr cells in ER+ human breast tumors is predictive of anti-estrogen treatment failure, presumably because these cells do not express the estrogen receptor, and thus would be selected for during long-term exposure to antiestrogen agents.41
In the mouse prostate, ALDHbr cells, as detected using the ALDEFLUOR™ kit, express high levels of Sca-1, a known marker of prostate in vivo engrafting cells.42,43 ALDHbr mouse prostate cells were the most efficient cells for generating prostatic outgrowths in an in vivo reconstitution assay. In the human prostate, ALDH1 expression is restricted to a subset of basal cells44, but the ALDEFLUOR™ assay did not selectively enrich for in vivo engrafting cells.45 In human prostate tumors, ALDH expression is correlated to Gleason scores and advanced-stage cancer, with high expression of the ALDH1A1 isoform associated with poor survival.44,46 As well, there is evidence that transplantation of ALDHbr cells are more tumorigenic and metastatic than ALDHlow cells.46,47
ALDH has also been reported to identify cells with stem cell properties in the normal colon.48 ALDH1-expressing cells are restricted to a subset of cells the bottom of the intestinal crypts, and these cells also express CD44 and CD133, both of which have been reported to be stem cell markers.49,50 The frequency of these ALDH-expressing cells increases as the normal tissue progresses to colon cancer, and ALDHbr are able to select for tumorigenic stem cells in some, but not all colon tumors.
The ALDEFLUOR™ assay has also been used to identify cancer stem-like/tumor-initiating cells in a wide variety of other types of malignancies, including pancreatic, liver, neural, pulmonary, thyroid, ovarian, head and neck, and gastric tumors.51–59
References
- Vasiliou V & Nebert DW. (2005) Hum Genomics 2(2): 138.
- Rice KL et al. (2008) Leuk Res 32(6): 873–83.
- Marcato P et al. (2011) Stem Cells 29(1): 32–45.
- Hartomo TB et al. (2015) Int J Oncol 46(3): 1089–98.
- Van Den Hoogen C et al. (2011) Clin Exp Metastasis 28(7): 615–25.
- Yagishita A et al. (2017) Bioconjug Chem 28(2): 302–6.
- Jones RJ et al. (1995) Blood 85(10): 2742–6.
- Storms RW et al. (1999) Proc Natl Acad Sci U S A 96(16): 9118–23.
- Fallon P & Gentry T. (2003) Br J … 122: 99–108.
- Hess DA et al. (2004) Blood 104(6): 1648–55.
- Storms RW et al. (2005) Blood 106(1): 95–102.
- Pearce DJ et al. (2005) Stem Cells 23(6): 752–60.
- Christ O et al. (2007) Haematologica 92(9): 1165–72.
- Roug AS et al. (2014) Cytotherapy 16(3): 392–401.
- Fatorova I et al. (2014) Biomed Res Int 2014.
- Lee HR et al. (2014) Transfusion 54(7): 1871–5.
- Shoulars K et al. (2016) Blood 127(19): 2346–54.
- Gerber JM et al. (2011) Am J Hematol 86(1): 31–7.
- Nakamura S et al. (2012) Int J Cancer 130(5): 1046–59.
- Cheung AMS et al. (2007) Leukemia 21(7): 1423–30.
- Ran D et al. (2009) Exp Hematol 37(12): 1423–1434.
- Gasparetto M et al. (2017) Haematologica 102(6): 1054–65.
- Gerber JM et al. (2012) Blood 119(15): 3571–7.
- Schuurhuis GJ et al. (2013) PLoS One 8(11).
- Muramoto GG et al. (2010) Stem Cells 28(3): 523–34.
- Pearce DJ & Bonnet D. (2007) Exp Hematol 35(9): 1437–46.
- Gasparetto M et al. (2012) Exp Hematol 40(10): 857–66.
- Ohta H et al. (2007) Exp Hematol 35(5): 817–30.
- Levi BP et al. (2009) Blood 113(8): 1670–80.
- Chang TH et al. (2014) Breast Cancer Res 16(1): R1.
- Tao L et al. (2015) Stem Cell Reports 5(1): 60–74.
- Shehata M et al. (2012) Breast Cancer Res 14(5): R134.
- Giraddi RR et al. (2015) Nat Commun 6: 8487.
- Rodilla V et al. (2015) PLoS Biol 13(2): e1002069.
- Wang C et al. (2017) Cell Rep 18(12): 2825–35.
- Van Keymeulen A et al. (2017) Cell Rep 20(7): 1525–32.
- Eirew P et al. (2012) Stem Cells 30(2): 344–8.
- Ginestier C et al. (2007) Cell Stem Cell 1(5): 555–67.
- Lim E et al. (2009) Nat Med 15(8): 907–13.
- Molyneux G et al. (2010) Cell Stem Cell 7(3): 403–17.
- Simoes BM et al. (2015) Cell Rep 12(12): 1968–77.
- Burger PE et al. (2005) Proc Natl Acad Sci U S A 102(20): 7180–5.
- Burger PE et al. (2009) Stem Cells 27(9): 2220–8.
- Le Magnen C et al. (2013) Clin Cancer Res 19(19): 5361–71.
- Gangavarapu KJ et al. (2013) Stem Cell Res Ther 4(5).
- Li T et al. (2010) Lab Investig 90(2): 234–44.
- Van Den Hoogen C et al. (2010) Cancer Res 70(12): 5163–73.
- Huang EH et al. (2009) Cancer Res 69(8): 3382–9.
- Dalerba P et al. (2007) Proc Natl Acad Sci U S A 104(24): 10158–63.
- O’Brien CA et al. (2007) Nature 445(7123): 106–10.
- Mullendore ME et al. (2009) Clin Cancer Res 15(7): 2291–301.
- Ma S et al. (2008) Mol Cancer Res 6(7): 1146–53.
- Bar EE et al. (2007) Stem Cells 25(10): 2524–33.
- Jiang F et al. (2009) Mol Cancer Res 7(3): 330–8.
- Hardin H et al. (2017) Lab Investig 97(10): 1142–51.
- Silva IA et al. (2011) Cancer Res 71(11): 3991–4001.
- Kryczek I et al. (2012) Int J Cancer 130(1): 29–39.
- Kulsum S et al. (2017) Mol Carcinog 56(2): 694–711.
- Wu D et al. (2016) Int J Oncol 49(2): 611–22.
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration

