STEMdiff™ APEL™ Medium
.
- Document # 28995
- Version 1.2.0
- Sep 2012
Background
Pluripotent stem cells (PSCs), including human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), are characterized by their ability to both self-renew and differentiate. PSCs have the potential to differentiate into cells from any somatic cell lineage, including those which emerge in development from the endodermal, mesodermal, and ectodermal germ layers.1
There are three main culture methods employed for directed differentiation: (1) co-culture with stromal cells or with conditioned media from stromal cell culture; (2) formation of embryoid bodies (EBs); and (3) adherent cell culture on extracellular-derived matrices. Co-culture methods involve the use of a uniquely supportive and inductive stromal cell type, which may or may not be derived from the target lineage, upon which the PSCs are placed or from which conditioned medium is extracted for later culture of PSCs. For example, hematopoietic differentiation can be induced by co-culture with M2-10B4 2 or OP93 stromal cells or by culture with conditioned medium from HepG2 stromal cells.4 This method is often quite efficient, but because unknown factors are secreted by the stromal cells, the mechanism of induction is poorly understood. EB-based differentiation involves removing hPSCs from adherent culture and placing them into non-adherent culture vessels, where they form spherical EBs and differentiate to a variety of lineages when subsequently plated onto a surface, such as gelatin. Lastly, emerging new protocols avoid EB formation, and instead keep the cells in adherent cell culture on extracellular-derived proteins or matrices. This method is therefore simpler to perform and can potentially be automated. At present, the EB-based and adherent cell-based methods both enable directed differentiation to a lineage of choice through the addition of specifi c factors. They are thus more amenable to experimental manipulation than stromal cell-based methods.
The factors used to induce differentiation to a given lineage have largely been identifi ed by studying the processes controlling normal embryogenesis,5 or by screening large numbers of molecules for their effect on lineage induction.6 As with embryogenesis, the factors need to be added in specific sequence, timing, and concentration, in order to induce fi rst the germ layer and then the specific lineage(s). For example, factors such as bone morphogenetic protein-4 (BMP-4) and Activin A might be added for 1 – 4 days, to induce hPSCs to the mesodermal germ layer;7-9 followed by a different combination of factors to induce those mesodermal cells to the cardiomyocyte8,10-12 or hematopoietic13,14 lineages. Similarly, defi nitive endoderm and subsequent pancreatic cell differentiation can be induced with a series of cytokine combinations including Activin A, FGF-10, cyclopamine, and retinoic acid.15,16 For each lineage, a growing list of factors have been shown to improve the efficiency of directed differentiation.
STEMdiff ™ APEL ™ Medium is a serum-free and animal component-free medium specifically developed to support hPSC differentiation. It was first described for the induction of hemato-endothelial cells, when supplemented with VEGF, BMP-4, SCF, and Activin A,7 but it has also been proven to be an effective basal medium for hPSC differentiation to other lineages, including cardiomyocytes.17 The following are examples of differentiation to cardiomyocyte, hematopoietic, or definitive endoderm cells from hPSCs using STEMdiff ™ APEL ™ Medium supplemented with cytokine combinations obtained from the literature. hPSCs can be differentiated either as EBs or in adherent cell-based systems, using STEMdiff ™ APEL ™ Medium supplemented with specific lineage-inducing factors. Moreover, the following examples were maintained in mTeSR ™1 feeder-independent medium for at least 10 passages prior to induction of differentiation. Using the right combination of growth factors, we expect that hPSCs can differentiate in STEMdiff ™ APEL ™ Medium to any somatic cell lineage, and we encourage users to try STEMdiff ™ APEL ™ Medium in other protocols as well.
Advantages of STEMdiff™ APEL™ Medium
- Defined
- Animal component-free
- Versatile, growth factor-free formulation to allow for differentiation to multiple lineages
For more details on the protocols used in these examples, contact STEMCELL Technologies Inc.’s Technical Support team at techsupport@stemcell.com.
Hematopoietic Differentiation
This protocol is based on Ng et al. 7 and Chadwick et al.,13 with the following changes: (1) STEMdiff ™ APEL ™ Medium was substituted for DMEM with 20% FBS as a basal medium; (2) cells were maintained prior to differentiation for at least 10 passages in mTeSR ™1 Maintenance Medium and Matrigel ™; (3) the entire differentiation procedure was performed in adherent cell culture, on a Matrigel ™coated surface instead of an EB-based method. Briefl y, hPSCs were passaged as clumps using dispase, onto Matrigel ™-coated 6-well plates in mTeSR ™1. Two days after passaging (Day 0), mTeSR ™1 medium was removed and replaced with 3 mL per well of STEMdiff ™ APEL ™ Medium supplemented with 30 ng/mL VEGF, 30 ng/mL BMP-4, 40 ng/mL SCF, and 50 ng/mL Activin A.7 On Day 4, this medium was removed and replaced with STEMdiff ™ APEL ™ Medium supplemented with 300 ng/mL SCF, 300 ng/mL Flt-3 ligand, 10 ng/mL IL-3, 10 ng/mL IL-6, 50 ng/mL G-CSF, 25 ng/mL BMP-4.13 On Days 7 and 10, complete media changes were done using STEMdiff ™ APEL ™ Medium and supplements as per Day 4. Finally on Day 13, cells were analyzed for hematopoietic differentiation by flow cytometry for expression of CD34 and CD45. Cells were also assayed by colony-forming unit (CFU) assay on Day 13, by plating 105 cells per dish in MethoCult ™ H4434 Classic, and counting the resultant colonies after 10 days. As shown in Figure 1, when STEMdiff ™ APEL ™ Medium was used with the published cytokine combinations in adherent cell based culture, approximately 30% of the cells expressed the hematopoietic marker CD34. Approximately 7% of the cells co-expressed CD34 and CD45, indicating the presence of hematopoietic progenitors (Figures 1A and 1B). This was further confirmed by the presence of CFUs at a frequency of approximately 1 in 1,000 cells (100 CFU colonies per 105 cells plated, Table 1). Interestingly, presumptive vascular endothelial cells, co-expressing CD34 and CD3115, 18 were also obtained by this procedure (Figure 1C). The frequencies of CD34+ cells, CD34+CD45+ cells, and CFUs shown here are in line with those in Chadwick et al.,13 and with more recent publications of hematopoietic differentiaton.14
This example demonstrates the utility of STEMdiff ™ APEL ™ Medium as a serum-free, animal component-free alternative for use in an established hematopoietic differentiation protocol.
Table 1. Emergence of hematopoietic colony forming units (CFUs)
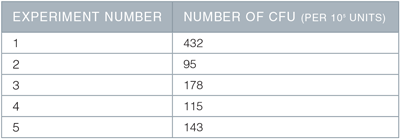
On Day 13 of the described protocol, 1 x 105 cells were plated into MethoCult ™ Classic H4434 for CFU assay, and the resulting colonies counted 10 days later. Shown are the number of CFU (colonies) per 1 x 105 cells plated, in 5 separate experiments, using 2 different hESC or hiPSC lines.
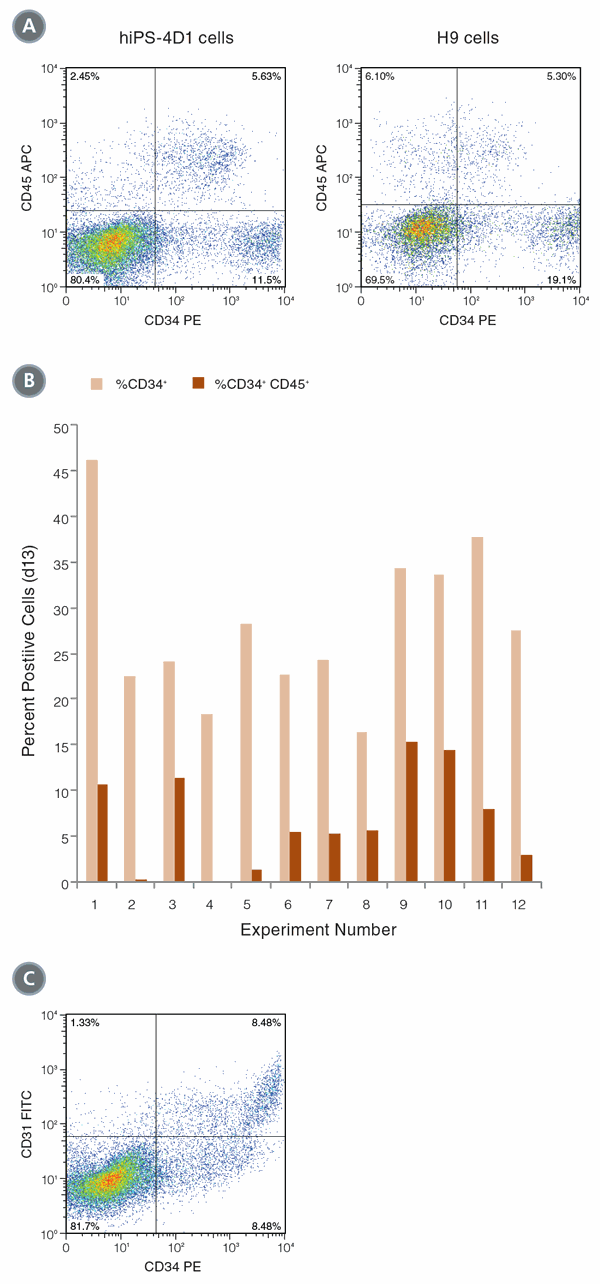
Figure 1. Differentiation of hPSCs into hematopoietic cells
(A) hiPS-4D1 cells (left) and H9 cells (right) were analyzed by flow cytometry on Day 13 of differentiation culture for expression of hematopoietic markers CD34 and CD45. (B) In 12 experiments, an average of 28.0% (± 8.6 STDEV standard deviation) of cells expressed CD34 on Day 13, and 7.3% (± 5.1%) of the cells were CD34+CD45+ double positive. (C) CD34hi cells co-expressed CD31, indicating that endothelial cells are also produced with this protocol.
Cardiomyocyte Differentiation
This protocol is based on Yang et al.,8 with the following changes: (1) STEMdiff ™ APEL ™ Medium was substituted for StemPro®-34 SFM as a basal medium; (2) cells were maintained prior to differentiation for at least 10 passages in mTeSR ™1 Maintenance Medium and Matrigel ™; (3) EBs were formed in AggreWell ™400 plates, and (4) the entire procedure was carried out within the AggreWell ™400 plate.
Briefl y, EBs of 1,000 cells each were formed in AggreWell ™400 plates, in STEMdiff ™ APEL ™ Medium supplemented with 0.5 ng/mL BMP-4 (Figure 2). After Day 1, media was changed to STEMdiff ™ APEL ™ Medium supplemented with 10 ng/mL BMP-4, 10 ng/mL Activin A, and 5 ng/mL bFGF. On Day 4, media was changed to STEMdiff ™ APEL ™ Medium supplemented with 10 ng/mL VEGF and 150 ng/mL Dkk1. On Day 8, media was changed again to STEMdiff ™ APEL ™ Medium supplemented with 10 ng/mL VEGF, 150 ng/mL Dkk1, and 10 ng/mL bFGF. On Day 12, fresh medium was replaced as per Day 8. On Day 16, the EBs were observed for synchronized beating, and cells were harvested for flow cytometry analysis of expression of the cardiomyocyte marker cardiac Troponin T (cTnT).
As shown in Figure 3, up to 95% of EBs were found to be beating on Day 16. Despite this high frequency of beating, the percentage of cells that were cTnT positive varied between 0.1% and 5.8%, which may indicate that a minority of cells within each EB were true cardiomyocytes. While this is lower than the frequency of cTnT+ cells in Yang et al.,8 it demonstrates that STEMdiff ™ APEL ™ Medium can support differentiation of mTeSR ™1-grown cells into beating cardiomyocytes with high efficiency. Further optimization will be required to increase the frequency of cTnT+ cells using this protocol. This example demonstrates the utility of STEMdiff ™ APEL ™ Medium as a serum-free, animal component-free alternative for use in an established cardiomyocyte differentiation protocol.
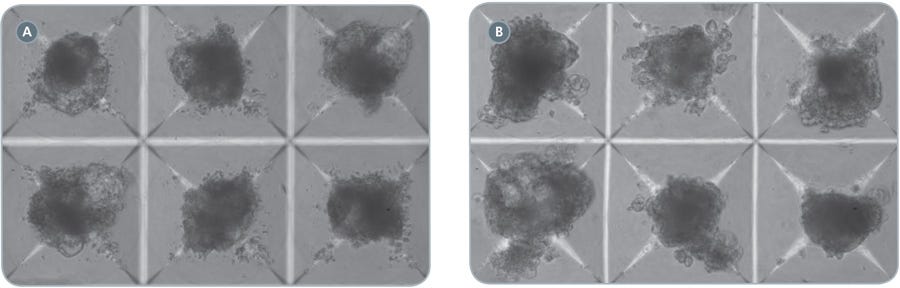
Figure 2. EBs in AggreWell ™400 cardiomyocyte differentiation protocol
hPSCs were differentiated into cardiomyocytes in STEMdiff ™APEL ™ Medium supplemented with cytokines as described. Shown are EBs in AggreWell ™400 plates, at Day 5 (A) or Day 13 (B) of the protocol. Transparent regions at the edges of the EBs tend to show beating. 100x magnification.
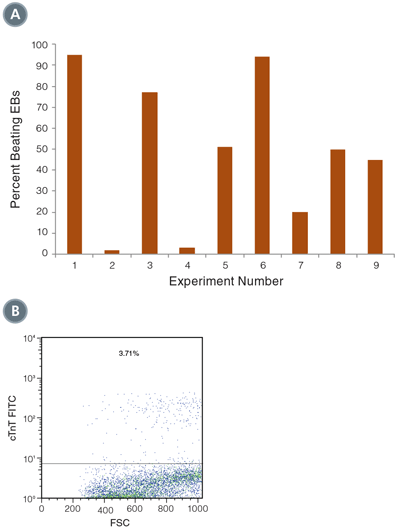
Figure 3. Differentiation of hPSCs into cardiomyocytes
(A) Beating EBs were counted on Day 16 of the culture, and varied between 5% and 95% of total EBs (n=9). (B) Expression of cTnT was measured on Day 16 of the culture by flow cytometry. Shown is a representative example of cTnT positive cells, from directed differentiation of H9 hESCs.
Definitive Endoderm Induction
This protocol is based on Rezania et al.,16 with the following changes: (1) STEMdiff ™ APEL ™ Medium was substituted for RMPI with 2% BSA as a basal medium; and (2) cells were maintained prior to the differentiation for at least 10 passages in mTeSR ™1 Maintenance Medium and Matrigel ™. Briefl y, PSCs were dissociated to single cells using ACCUTASE ™, and 3x106 cells per well were plated onto Matrigel ™-coated 6-well plates, in 2 mL STEMdiff ™ APEL ™ Medium supplemented with 10 μM Y-27632 Rock Inhibitor. On Day 1, media was removed and replaced with STEMdiff ™ APEL ™ Medium supplemented with 100 ng/mL Activin A, 20 ng/mL Wnt3a, and 8 ng/mL bFGF. Media was replaced daily with STEMdiff ™ APEL ™ Medium, on Days 2 and 3. On Day 4, cells were harvested and analyzed by flow cytometry for expression of definitive endodermal markers, CXCR4 and SOX-17. As shown in Figure 4, the majority of cells co-expressed CXCR4 and SOX-17 after 4 days in STEMdiff ™ APEL ™ Medium supplemented with endoderm-inducing cytokines. An average of 69.53 ± 9.1% CXCR4+SOX-17+ double positive cells were obtained using this procedure (n=2, each in triplicate). The frequency of cells expressing CXCR4 (>90%) is consistent with data shown in Rezania et al.16 indicating that STEMdiff ™ APEL ™ Medium can successfully generate definitive endoderm from hESCs cultured in mTeSR ™1.
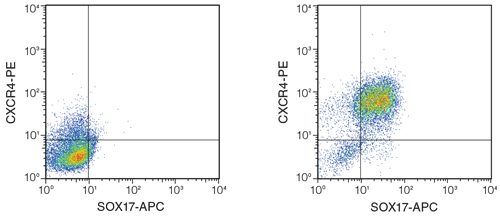
Figure 4. Differentiation of hPSCs into definitive endoderm using STEMdiff ™ APEL ™ Medium and published cytokines.
H9 hESCs were differentiated based on Rezania et al.16 On Day 4, cells were highly positive for CXCR4 and SOX-17, which together mark definitive endoderm. Left: cells cultured in STEMdiff ™ APEL ™ Medium alone (no cytokines added); Right: differentiated cells after culture in STEMdiff ™ APEL ™ Medium with inducing cytokines.
Summary
STEMdiff ™ APEL ™ Medium is a serum-free, animal component-free basal medium which, when supplemented with specific cytokines, supports the directed differentiation of human PSCs. The examples shown here demonstrate its versatility for hPSC differentiation to cardiomyocyte, hematopoietic, or definitive endodermal lineages. Other lineages may be possible, with optimized combination, concentrations, and timing of addition of cytokines or small molecule inhibitors.
Optimization of the above protocols may be required, as different cell lines may respond differently to a given stimulus. The quality of the starting population is critical, and starting cultures should have little morphologically identifiable differentiated cells (less than 15%). Cytokines should be titrated due to batch variation and for their specific activity on a given cell line.
References
- Itskovitz-Eldor J et al. Mol Med 6:88-95, 2000
- Hill KL et al. Current Protocols in Stem Cell Biology, 2007
- Woods N-B et al. Stem Cells. 2011 May 4. doi: 10.1002/stem.657
- Lu M et al. Experimental Hematology 37:924-936.e924, 2009
- Gadue P, et al. Experimental Hematology 33:955-964, 2005
- Borowiak M et al. Cell Stem Cell 4:348-358, 2009
- Ng ES et al. Nat. Protocols 3:768-776, 2008
- Yang L et al. Nature 453:524-528, 2008
- Burridge PW et al. PLoS ONE 6:e18293, 2011
- Evseenko D et al. PNAS 107:13742-13747, 2010
- Filipczyk A et al. Cellular and Molecular Life Sciences 64:704-718, 2007
- Gaur M et al. Cytotherapy 12:807-817, 2010
- Chadwick K et al. Blood 102:906-915, 2003
- Chicha L et al. PLoS ONE 6:e14733, 2011
- D’Amour KA et al. Nat Biotech 24:1392-1401, 2006
- Rezania A, et al. Diabetes 60:239-247, 2011
- Elliott DA et al. Nat Methods 8(12):1037-1040, 2011
- Choi K-D et al. Stem Cells 27:559-567, 2009
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration


