Enhance Sensitivity of Multiple Myeloma Testing with Purified CD138+ Plasma Cells
.
- Document # 28731
- Version 2.0.0
- Nov 2019
Multiple myeloma bone marrow samples typically comprise a mix of non-malignant and malignant cells. It can be challenging to detect the small number of malignant myeloma cells when analyzing samples with a large proportion of non-malignant cells. Purification of plasma cells by CD138 positive selection increases the proportion of malignant myeloma cells in the analyte and thereby enhances the detection of genomic aberrations in the sample.
This technical bulletin presents a protocol for the isolation of plasma cells to enhance the sensitivity of downstream analysis such as fluorescence in situ hybridization (FISH), microarray-based assays, genomic sequencing, and gene expression profiling for multiple myeloma testing.
Background
Multiple Myeloma
Multiple myeloma is a form of cancer caused by B cell neoplasia that results in dysregulated production and clonal expansion of malignant plasma cells (cells that express CD138 (Syndecan-1) and are involved in the production of antibodies during an immune response).1 The disease is characterized by excessive numbers of abnormal plasma cells in the bone marrow and overproduction of both intact monoclonal immunoglobulins and free monoclonal kappa and lambda immunoglobulin light chains.
Genomic Aberration Analysis of Multiple Myeloma Samples
Detection and quantification of CD138+ plasma cells in bone marrow is typically the first laboratory screening method for the disease. Multiple myeloma can be distinguished from other B cell neoplasias by the presence of characteristic chromosome aberrations. These chromosome aberrations include chromosome translocation events in the immunoglobulin heavy chain region that result in oncogene activations, chromosomal deletions (particularly of chromosome 13), and hyperploidy. This information can also be used to classify multiple myeloma into subtypes with different characteristics.2, 3
Molecular cytogenetics techniques such as FISH are widely used tools for characterizing multiple myeloma. FISH employs nucleic acid probes to detect and localize the presence or absence of specific DNA sequences on chromosomes. The FISH assay is limited by the ability of the existing nucleic acid probes to detect genomic aberrations. Therefore, other techniques with higher resolution (e.g. microarray-based genomic profiling assays, genomic sequencing, and gene expression profiling) are being explored as additional tests to be used in combination with FISH for multiple myeloma testing.4,5,6
Enhanced Sensitivity with CD138+ Plasma Cell Enrichment
Multiple myeloma bone marrow samples typically comprise a mix of nonmalignant and malignant cells. It can be challenging to detect the small number of malignant plasma cells within the large proportion of healthy B cells and non-malignant plasma cells that exhibit a normal karyotype. To identify and analyze the small fraction of abnormal clones it would be necessary to analyze a large number of cells. However, the CD138 antigen is present on all plasma cells (both non-malignant and malignant cells) but not on mature B cells. This makes the CD138 antigen a suitable selection marker for the enrichment of all plasma cells including the malignant multiple myeloma cells. Plasma cell enrichment by CD138 isolation thereby increases the proportion of malignant myeloma cells in the analyte and enhances sensitivity in detecting genomic aberrations compared to analyzing samples with mixed cell populations.4,5,7 Plasma cell enrichment by CD138 selection is therefore a beneficial step prior to downstream analysis to enhance sensitivity and obtain more reliable FISH, microarraybased genomic profiling, and genomic sequencing testing for multiple myeloma.
The National Comprehensive Cancer Network® (NCCN Guidelines Version 2.2020 Multiple Myeloma) recommends the use of enriched plasma cells obtained from bone marrow aspirations when performing FISH analysis for multiple myeloma testing.
- NCCN Guidelines® Version 2.2020 Multiple Myeloma; NCCN.org
Easy, Column-Free Plasma Cell Enrichment
One method to obtain CD138+ plasma cells from bone marrow samples or peripheral blood mononuclear cells is by using an immunomagnetic cell isolation technology such as EasySep™ (see Figure 1).

With EasySep™, the desired cells are targeted using antibody complexes recognizing CD138 on the surface of the cell and linking the cells to magnetic particles. The sample is then placed in an EasySep™ magnet, the labeled cells are pulled to the side of the tube, and the unlabeled cells can be simply poured or pipetted off into a new tube.
The enrichment of CD138+ cells with EasySep™ has been shown to be an effective method to obtain plasma cells for downstream multiple myeloma testing.5,6,8 For increased time-savings and minimized sample handling, plasma cell enrichment with EasySep™ can be fully automated with the RoboSep™ instruments.5,8 A study to evaluate RoboSep™ for CD138+ plasma cell enrichment found that the instrument reliably sorted plasma cells even when they were found in low frequencies ( <2%).8
The following section provides a complete protocol for plasma cell enrichment from bone marrow, whole blood, and mononuclear cell preparations for subsequent downstream multiple myeloma testing.
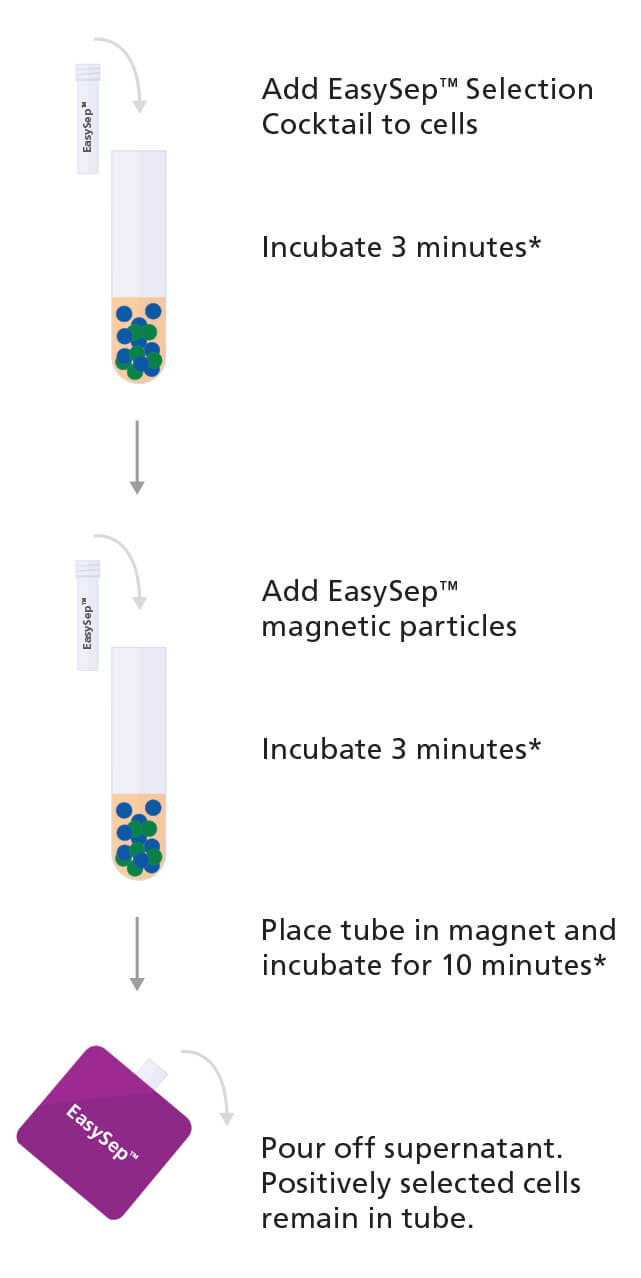
Figure 1. Sample Protocol for Manual Selection of CD138+ Cells Using EasySep™ Positive Selection Kit
CD138+ cells are labeled with antibodies and magnetic particles and separated using an EasySep™ magnet. Isolated cells are immediately available for downstream applications such as fluorescence in situ hybridization (FISH), flow cytometry, culture, or DNA/RNA extraction.
*Times will vary depending on the specific reagent, the isolation protocol, and separation platform.
Protocol
PART 1: Source-Dependent Sample Preparation
Sample Source: Bone Marrow
Sample Preparation with EasySep™ Human Whole Blood and Bone Marrow CD138 Positive Selection Kit II - Catalog #17887Dilute the sample 5- to 10- fold in D-PBS (without calcium and magnesium) and mix gently by pipetting up and down. Filter the sample through a pre-wetted 70 - 100 μm strainer to remove bone fragments, cell aggregates, and debris. Transfer the bone marrow single cell suspension into a tube and centrifuge at 300 x g for 10 minutes with the brake off. Carefully remove and discard the plasma, without disturbing the buffy coat/red blood cell pellet, and resuspend to the original sample volume with D-PBS. For samples more than 24 hours old, add DNase I Solution at 100 μg/mL to help reduce cell clumping. DNase I Solution can be added directly to pelleted cells, with gentle mixing, before resuspension.
Transfer a maximum of 4.5 mL of the single cell suspension into a round bottom 14 mL polystyrene tube. Add 1X EasySep™ Red Blood Cell Lysis Buffer* at a ratio of 1 part lysis buffer to 1 part sample and mix well. The sample is now ready for manual (Part 2A) or automated (Part 2B) cell separation.
Sample Preparation with EasySep™ Human CD138 Positive
Selection Kit II - Catalog #17877
Prepare a mononuclear cell (MNC) suspension from whole bone
marrow by centrifugation over a density gradient medium (e.g.
Lymphoprep™). Alternately, remove red blood cells
by lysis using Ammonium Chloride Solution. After
preparation, resuspend cells at 1 x 108 cells/mL in in PBS containing 2%
fetal bovine serum and 1 mM EDTA and transfer a maximum of 8.5 mL
to a round bottom 14 mL polystyrene tube. The
sample is now ready for manual (Part 2A) or automated (Part 2B) cell
separation.
Sample Source: Peripheral Blood
Sample Preparation with EasySep™ Human Whole Blood and Bone
Marrow CD138 Positive Selection Kit II - Catalog #17887
Collect whole blood in a blood collection tube containing
anticoagulant. Transfer a maximum of 4.5 mL whole blood to around bottom 14 mL polystyrene tube. Add 1X EasySep™ Red Blood Cell Lysis Buffer* at a ratio of 1 part lysis buffer to 1 part
blood sample and mix well. The sample is now ready for manual (Part
2A) or automated (Part 2B) separation.
Sample Preparation with EasySep™ Human CD138 Positive
Selection Kit II - Catalog #17877
Collect whole blood in a blood collection tube containing anticoagulant. Prepare a peripheral blood mononuclear cell (PBMC) suspension from whole blood by centrifugation over a density gradient medium (e.g. Lymphoprep™).
After preparation, resuspend cells at 1 x 108 cells/mL in PBS containing 2% fetal bovine serum and 1 mM EDTA and transfer a maximum of 8.5 mL to a round bottom 14 mL polystyrene tube. The sample is now ready for manual (Part 2A) or automated (Part 2B) cell separation.
Sample Source: Frozen Bone Marrow or Peripheral Blood Mononuclear Cells
Sample Preparation with EasySep™ Human CD138 Positive Selection Kit II - Catalog #17877Incubate the cells with DNase I Solution at a concentration of 100 μg/mL at room temperature (15 - 25°C) for at least 15 minutes. Filter aggregated suspensions through a 37 μm strainer for optimal results. After preparation, resuspend cells at 1 x 108 cells/mL in PBS containing 2% fetal bovine serum and 1 mM EDTA. Transfer a maximum of 8.5 mL to a round bottom 14 mL polystyrene tube. The sample is now ready for manual (Part 2A) or automated (Part 2B) cell separation.
*Note: EasySep™ Red Blood Cell Lysis Buffer is supplied as a 10X concentrate in the kit. Prepare 1X lysis buffer at least 1 hour before use by adding 1 part 10X lysis buffer to 9 parts distilled or Type 1 water. Mix gently and completely before use.
PART 2A: Manual Plasma Cell Enrichment
Follow the specific instructions on the corresponding product information sheet for the labeling of cells with the EasySep™ Selection Cocktail and RapidSpheres™ (Figure 1). For the first step, add the EasySep™ Selection Cocktail and RapidSpheres™ to label the desired cells with an antibody complex against CD138+ (Syndecan-1) and magnetic particles. Next, place the sample tube inside the EasySep™ magnet, incubate and then pipette or pour off the supernatant. The desired CD138+ cells will remain in the tube and the unwanted cells will have been poured or pipetted off. Remove the tube containing the desired cells from the magnet and resuspend cells in the appropriate medium. The isolated cells will contain EasySep™ magnetic particles attached to the surface. However these do not do not interfere with downstream applications such as flow cytometry, FISH and nucleic acid isolation.
For more information, use the product information sheet (PIS) finder to search and download the specific PIS for your product catalog number.
PART 2B: Automated Plasma Cell Enrichment
The cell isolation step to enrich for plasma cells can be fully automated using the RoboSep™-S and RoboSep™-16 instruments. The instruments perform all cell labeling and separation steps. To use RoboSep™ for CD138+ plasma cell enrichment, select the optimized instrument protocol (detailed in the corresponding Product Information Sheet), load the samples and reagents and start the cell separation process by following the on-screen prompts. When the run is complete remove the tube containing the isolated cells and resuspend in desired medium. Be sure to collect cells from the sides of the tube.

Why Use RoboSep™?
Streamline and standardize your cell isolations to minimize hands-on time and increase laboratory throughput with the fully automated RoboSep™ instruments. Isolated cells are immediately ready for downstream applications such as fluorescence in situ hybridization (FISH), flow cytometry, culture, or DNA/RNA extraction.
PART 3: Assessing Cell Purity After Cell Isolation
For purity assessment of CD138+ cells by flow cytometry use Anti-Human CD138 (Syndecan-1) Antibody, Clone MI15. Plasma cells express either the κ (Kappa) or λ (Lambda) light chain and cell purity can also be assessed by staining for intracellular κ and λ light chains as described by Ahmann and colleagues.9 Alternatively you can use markers such as Anti-Human CD38 Antibody, Clone HIT2 and Anti-Human CD45 Antibody, Anti-Human CD45 Antibody, Clone HI30 to detect CD38+CD45 variable cells.10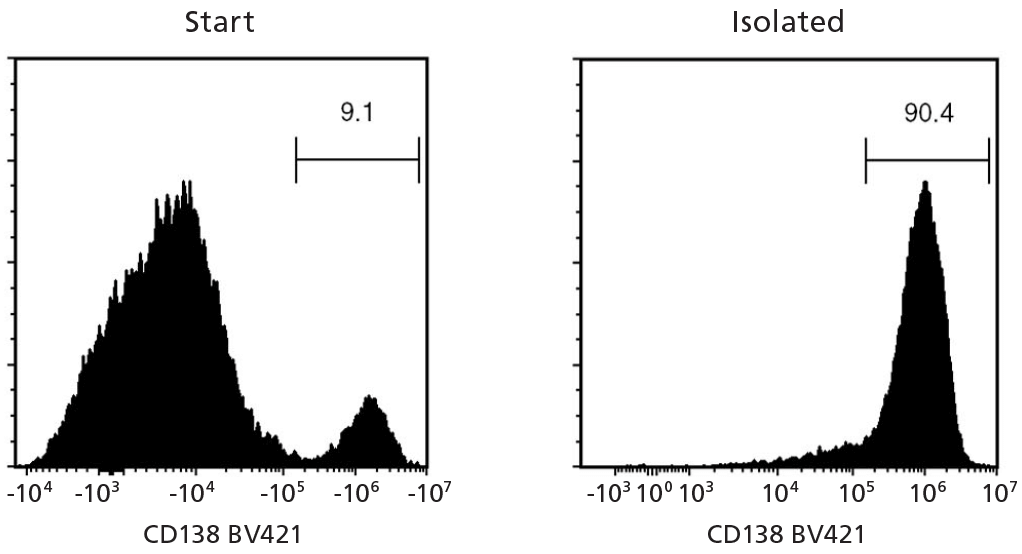
Figure 2. EasySep™ Human Whole Blood and Bone Marrow CD138 Positive Selection Kit II (Catalog #17887)
Starting with fresh whole blood spiked with a multiple myeloma cell line, U266, the CD138+ cell content of the selected fraction typically ranges from 83.7 - 98.3%. In the above example, the purities of the start and final isolated fractions are 9.1% and 90.4%, respectively.
NOTE: Red blood cells were removed from the start sample by lysis prior to flow cytometry. For samples with CD138+ starting frequency < 10 - 15%, the CD138+ purity of the isolated fraction may be variable.
PART 4: Downstream Testing
Enriched plasma cells are ready for downstream testing and analysis using techniques such as FISH, microarray based genomic profiling, and genomic sequencing.
Product Listing
Cell Separation Products
RoboSep™ Instruments
RoboSep™ instruments fit easily into the workflow of any lab that needs the multi-sample processing capacity, speed, reliability, and convenience of automated cell isolation.
Try RoboSep™ in your lab to see how it can improve sample processing efficiency!
Staining Antibodies for Purity Assessment by Flow Cytometry
References
- Huff CA & Matsui W. (2008) Multiple myeloma cancer stem cells. J Clin Oncol 26(17): 2895–900.
- Zandecki M et al. (1996) Multiple myeloma: almost all patients are cytogenetically abnormal. Br J Haematol 94(2): 217–27.
- Facon T et al. (2001) Chromosome 13 abnormalities identified by FISH analysis and serum beta2-microglobulin produce a powerful myeloma staging system for patients receiving high-dose therapy. Blood 97(6): 1566–71.
- Pugh TJ et al. (2018) Assessing genome-wide copy number aberrations and copy-neutral loss-of-heterozygosity as best practice: An evidencebased review from the Cancer Genomics Consortium working group for plasma cell disorders. Cancer Genet 228–229: 184–196.
- Kjeldsen E. (2016) Identification of Prognostically Relevant Chromosomal Abnormalities in Routine Diagnostics of Multiple Myeloma Using Genomic Profiling. Cancer Genomics Proteomics 13(2): 91–127.
- Berry NK et al. (2014) Genomic profiling of plasma cell disorders in a clinical setting: integration of microarray and FISH, after CD138 selection of bone marrow. J Clin Pathol 67(1): 66–9.
- Glaskova L et al. (2017) CD138 Enrichment Strategy and Results of Chromosome Genomic Array Testing (CGAT) for Multiple Myelomas. Cancer Genet 214: 43.
- Shetty S et al. (2012) Utility of a column-free cell sorting system for separation of plasma cells in multiple myeloma FISH testing in clinical laboratories. Int J Hematol 95(3): 274–81.
- Ahmann GJ et al. (1998) A novel three-color, clone-specific fluorescence in situ hybridization procedure for monoclonal gammopathies. Cancer Genet Cytogenet 101(1): 7–11.
- Kumar S et al. (2010) Immunophenotyping in multiple myeloma and related plasma cell disorders. Best Pract Res Clin Haematol 23(3): 433–51.
- NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®): Multiple Myeloma [Online], Version 2.2020. [https://www.nccn.org/professionals/physician_gls/recently_updated.aspx] (accessed Oct 23, 2019)
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration



